Abstract
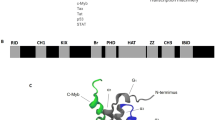
This study aims to gain insight into the DNA-specific recognition mechanism of c-Myb transcription factor during the regulation of cell early differentiation and proliferation. Therefore, we chose the chicken myeloid gene, mitochondrial import protein 1 (mim-1), as a target to study the binding specificity between potential dual-Myb-binding sites. The c-Myb-binding site in mim-1 is a pseudo-palindromic sequence AACGGTT, which contains two AACNG consensuses. Simulation studies in different biological scenarios revealed that c-Myb binding with mim-1 in the forward strand (complex F) is more stable than that in the reverse strand (complex R). The principal component analysis (PCA) dynamics trajectory analyses suggested an opening motion of the recognition helices of R2 and R3 (R2R3), resulting in the dissociation of DNA from c-Myb in complex R at 330 K, triggered by the reduced electrostatic potential on the surface of R2R3. Furthermore, the DNA confirmation and hydrogen-bond interaction analyses indicated that the major groove width of DNA increased in complex R, which affected on the hydrogen-bond formation ability between R2R3 and DNA, and directly resulted in the dissociation of DNA from R2R3. The steered molecular dynamics (SMD) simulation studies also suggested that the electrostatic potential, major groove width, and hydrogen bonds made major contribution to the DNA-specific recognition. In vitro trials confirmed the simulation results that c-Myb specifically bound to mim-1 in the forward strand. This study indicates that the three-dimensional (3D) structure features play an important role in the DNA-specific recognition mechanism by c-Myb besides the AACNG consensuses, which is beneficial to understanding the cell early differentiation and proliferation regulated by c-Myb, as well as the prediction of novel c-Myb-binding motifs in tumorigenesis.
摘要
本研究旨在探索c-Myb转录因子在调控细胞早期分化和增殖过程中的DNA特异性识别机制. 我们以鸡髓系基因mim-1为研究对象, 研究其潜在双c-Myb结合位点的结合特异性. mim-1的c-Myb结合位点是一个伪回文序列AACGGTT, 正反方向分别包含1个c-Myb结合保守序列AACNG. 不同条件下的重复分子动力学模拟研究表明, c-Myb与mim-1正链的结合(复合物F)比与反链的结合(复合物R)更稳定. 主成分分析(PCA)动力学轨迹分析表明, 在330 K温度下, c-Myb识别螺旋R2和R3(R2R3)的开放运动导致复合物R中DNA与c-Myb解离, 且此开放运动是由R2R3表面静电势降低引起. 同时, DNA构象和氢键相互作用分析表明, 复合物R中DNA的大沟宽度增加影响了R2R3与DNA间的氢键形成, 直接导致DNA与R2R3的解离. 拉伸分子动力学模拟研究进一步表明, 静电势、 DNA大沟宽度和氢键对DNA特异性识别起到重要作用. 体外实验证实了计算模拟结果, 即c-Myb只与mim-1正链结合. 本研究表明, 除一维保守序列AACNG外, 三维结构特性对c-Myb的DNA特异性识别也起着重要作用. 本研究结果有助于理解c-Myb在细胞早期分化和增殖中的调控机制, 以及预测开发由c-Myb结合位点增多引起的肿瘤发生标志物.
Similar content being viewed by others
Data availability statement
The structure of c-Myb binding to DNA of MBS1 is available on the Protein Data Bank (PDB code: No. 1MSE).
Change history
13 April 2024
An Erratum to this paper has been published: https://doi.org/10.1631/jzus.B22e0634
References
Abaza MSI, Al-Attiyah RJ, Al-Saffar AM, et al., 2004. Anti-sense oligodeoxynucleotide directed against c-myb has anticancer activity and potentiates the antiproliferative effect of conventional anticancer drugs acting by different mechanisms in human colorectal cancer cells. Tumor Biol, 24(5):241–257. https://doi.org/10.1159/000076139
Aggarwal P, Bhavesh NS, 2021. Hinge like domain motion facilitates human RBMS1 protein binding to proto-oncogene c-myc promoter. Nucleic Acids Res, 49(10):5943–5955. https://doi.org/10.1093/nar/gkab363
Berendsen HJC, van der Spoel D, van Drunen R, 1995. GROMACS: a message-passing parallel molecular dynamics implementation. Comp Phys Comm, 91(1–3):43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Bernstein FC, Koetzle TF, Williams GJB, et al., 1978. The protein data bank: a computer-based archival file for macromolecular structures. Arch Biochem Biophys, 185(2): 584–591. https://doi.org/10.1016/0003-9861(78)90204-7
Bhattarai G, Lee YH, Lee MH, et al., 2013. Gene delivery of c-myb increases bone formation surrounding oral implants. J Dent Res, 92(9):840–845. https://doi.org/10.1177/0022034513497753
Biedenkapp H, Borgmeyer U, Sippel AE, et al., 1988. Viral myb oncogene encodes a sequence-specific DNA-binding activity. Nature, 335(6193):835–837.
Cicirò Y, Sala A, 2021. MYB oncoproteins: emerging players and potential therapeutic targets in human cancer. Oncogenesis, 10(2):19. https://doi.org/10.1038/s41389-021-00309-y
Clesham K, Walf-Vorderwülbecke V, Gasparoli L, et al., 2022. Identification of a c-MYB-directed therapeutic for acute myeloid leukemia. Leukemia, 36(6):1541–1549. https://doi.org/10.1038/s41375-022-01554-9
Dai LQ, Yu J, 2020. Inchworm stepping of Myc-Max heterodimer protein diffusion along DNA. Biochem Biophys Res Commun, 533(1):97–103. https://doi.org/10.1016/j.bbrc.2020.08.004
Essmann U, Perera L, Berkowitz ML, et al., 1995. A smooth particle mesh Ewald method. J Chem Phys, 103(19): 8577–8593. https://doi.org/10.1063/1.470117
Fuglerud BM, Lemma RB, Wanichawan P, et al., 2017. A c-Myb mutant causes deregulated differentiation due to impaired histone binding and abrogated pioneer factor function. Nucleic Acids Res, 45(13):7681–7696. https://doi.org/10.1093/nar/gkx364
Gautam S, Fioravanti J, Zhu W, et al., 2019. The transcription factor c-Myb regulates CD8+ T cell stemness and anti-tumor immunity. Nat Immunol, 20(3):337–349. https://doi.org/10.1038/s41590-018-0311-z
Gonda TJ, Leo P, Ramsay RG, 2008. Estrogen and MYB in breast cancer: potential for new therapies. Expert Opin Biol Ther, 8(6):713–717. https://doi.org/10.1517/14712598.8.6.713
Graf T, 1992. Myb: a transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr Opin Genet Dev, 2(2):249–255. https://doi.org/10.1016/S0959-437X(05)80281-3
Greig KT, de Graaf CA, Murphy JM, et al., 2010. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood, 115(14):2796–2805. https://doi.org/10.1182/blood-2009-08-239210
Hao DY, Ohme-Takagi M, Sarai A, 1998. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. J Biol Chem, 273(41):26857–26861. https://doi.org/10.1074/jbc.273.41.26857
Hao DY, Yamasaki K, Sarai A, et al., 2002. Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry, 41(13):4202–4208. https://doi.org/10.1021/bi015979v
Hess B, Bekker H, Berendsen HJC, et al., 1997. LINCS: a linear constraint solver for molecular simulations. J Comput Chem, 18(12):1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H
Heuser C, Gattinoni L, 2022. c-Myb redefines the hierarchy of stem-like T cells. Nat Immunol, 23(10):1405–1407. https://doi.org/10.1038/s41590-022-01319-7
Hugo H, Cures A, Suraweera N, et al., 2006. Mutations in the MYB intron I regulatory sequence increase transcription in colon cancers. Genes Chromosomes Cancer, 45(12): 1143–1154. https://doi.org/10.1002/gcc.20378
Kanei-Ishii C, Yasukawa T, Morimoto RI, et al., 1994. c-Myb-induced trans-activation mediated by heat shock elements without sequence-specific DNA binding of c-Myb. J Biol Chem, 269(22):15768–15775. https://doi.org/10.1016/S0021-9258(17)40747-2
Katzen AL, Kornberg TB, Bishop JM, 1985. Isolation of the proto-oncogene c-myb from D. melanogaster. Cell, 41(2): 449–456. https://doi.org/10.1016/S0092-8674(85)80018-0
Kawasaki M, Oda M, 2021. DNA-binding function of c-Myb R2R3 around thermal denaturation temperature. Biophys Physicobiol, 18:78–84. https://doi.org/10.2142/biophysico.bppb-v18.009
Krissinel E, Henrick K, 2007. Inference of macromolecular assemblies from crystalline state. J Mol Biol, 372(3):774–797. https://doi.org/10.1016/j.jmb.2007.05.022
Lauria A, Ippolito M, Almerico AM, 2009. Principal component analysis on molecular descriptors as an alternative point of view in the search of new Hsp90 inhibitors. Comput Biol Chem, 33(5):386–390. https://doi.org/10.1016/j.compbiolchem.2009.07.010
Lavery R, Moakher M, Maddocks JH, et al., 2009. Conformational analysis of nucleic acids revisited: Curves+. Nucleic Acids Res, 37(17):5917–5929. https://doi.org/10.1093/nar/gkp608
Lee YH, Kim HS, Kim JS, et al., 2016. C-myb regulates autophagy for pulp vitality in glucose oxidative stress. J Dent Res, 95(4):430–438. https://doi.org/10.1177/0022034515622139
Lindorff-Larsen K, Piana S, Palmo K, et al., 2010. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins, 78(8):1950–1958. https://doi.org/10.1002/prot.22711
Lu H, Wang Y, Huang Y, et al., 2013. Expression and prognostic role of c-Myb as a novel cell cycle protein in esophageal squamous cell carcinoma. Clin Transl Oncol, 15(10):796–801. https://doi.org/10.1007/s12094-013-1009-1
Madan A, Radha PK, Hosur RV, et al., 1995. Bacterial expression, characterization and DNA binding studies on Drosophila melanogaster c-Myb DNA-binding protein. Eur J Biochem, 232(1):150–158. https://doi.org/10.1111/j.1432-1033.1995.tb20793.x
Malaterre J, Mantamadiotis T, Dworkin S, et al., 2008. c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells, 26(1):173–181. https://doi.org/10.1634/stemcells.2007-0293
Mansour MR, Abraham BJ, Anders L, et al., 2014. Oncogene regulation. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science, 346(6215):1373–1377. https://doi.org/10.1126/science.1259037
Maurice D, Hooper J, Lang G, et al., 2007. c-myb regulates lineage choice in developing thymocytes via its target gene Gata3. EMBO J, 26(15):3629–3640. https://doi.org/10.1038/sj.emboj.7601801
Mitani Y, Li J, Rao PH, et al., 2010. Comprehensive analysis of the MYB-NFIB gene fusion in salivary adenoid cystic carcinoma: incidence, variability, and clinicopathologic significance. Clin Cancer Res, 16(19):4722–4731. https://doi.org/10.1158/1078-0432.CCR-10-0463
Miyazaki T, Pan Y, Joshi K, et al., 2012. Telomestatin impairs glioma stem cell survival and growth through the disruption of telomeric G-quadruplex and inhibition of the protooncogene, c-Myb. Clin Cancer Res, 18(5):1268–1280. https://doi.org/10.1158/1078-0432.CCR-11-1795
Morris GM, Huey R, Lindstrom W, et al., 2009. AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem, 30(16):2785–2791. https://doi.org/10.1002/jcc.21256
Ness SA, 1996. The Myb oncoprotein: regulating a regulator. Biochim Biophys Acta, 1288(3):F123–F139. https://doi.org/10.1016/s0304-419x(96)00027-3
Ness SA, Marknell Å, Graf T, 1989. The v-myb oncogene product binds to and activates the promyelocyte-specific mim-1 gene. Cell, 59(6):1115–1125. https://doi.org/10.1016/0092-8674(89)90767-8
Ni SJ, Zhu JY, Zhang JG, et al., 2015. Expression and clinical role of NF45 as a novel cell cycle protein in esophageal squamous cell carcinoma (ESCC). Tumor Biol, 36(2): 747–756. https://doi.org/10.1007/s13277-014-2683-5
Nishina Y, Nakagoshi H, Imamoto F, et al., 1989. Trans-activation by the c-myb proto-oncogene. Nucleic Acids Res, 17(1):107–117. https://doi.org/10.1093/nar/17.1.107
O’Boyle NM, Banck M, James CA, et al., 2011. Open Babel: an open chemical toolbox. J Cheminform, 3:33. https://doi.org/10.1186/1758-2946-3-33
Ogata K, Hojo H, Aimoto S, et al., 1992. Solution structure of a DNA-binding unit of Myb: a helix-turn-helix-related motif with conserved tryptophans forming a hydrophobic core. Proc Natl Acad Sci USA, 89(14):6428–6432. https://doi.org/10.1073/pnas.89.14.6428
Ogata K, Morikawa S, Nakamura H, et al., 1994. Solution structure of a specific DNA complex of the Myb DNA-binding domain with cooperative recognition helices. Cell, 79(4):639–648. https://doi.org/10.1016/0092-8674(94)90549-5
Ogata K, Morikawa S, Nakamura H, et al., 1995. Comparison of the free and DNA-complexed forms of the DMA-binding domain from c-Myb. Nat Struct Biol, 2(4):309–320. https://doi.org/10.1038/nsb0495-309
Ogata K, Kanei-Ishii C, Sasaki M, et al., 1996. The cavity in the hydrophobic core of Myb DNA-binding domain is reserved for DNA recognition and trans-activation. Nat Struct Biol, 3(2):178–187. https://doi.org/10.1038/nsb0296-178
Oh IH, Reddy EP, 1999. The myb gene family in cell growth, differentiation and apoptosis. Oncogene, 18(19):3017–3033. https://doi.org/10.1038/sj.onc.1202839
Ording E, KvÅvik W, Bostad A, et al., 1994. Two functionally distinct half sites in the DNA-recognition sequence of the Myb oncoprotein. Eur J Biochem, 222(1):113–120. https://doi.org/10.1111/j.1432-1033.1994.tb18848.x
Padroni G, Withers JM, Taladriz-Sender A, et al., 2019. Sequence-selective minor groove recognition of a DNA duplex containing synthetic genetic components. J Am Chem Soc, 141(24):9555–9563. https://doi.org/10.1021/jacs.8b12444
Pan JH, Adair-Kirk TL, Patel AC, et al., 2014. Myb permits multilineage airway epithelial cell differentiation. Stem Cells, 32(12):3245–3256. https://doi.org/10.1002/stem.1814
Pan YP, Nussinov R, 2007. Structural basis for p53 binding-induced DNA bending. J Biol Chem, 282(1):691–699. https://doi.org/10.1074/jbc.M605908200
Pardo-Saganta A, Law BM, Tata PR, et al., 2015. Injury induces direct lineage segregation of functionally distinct airway basal stem/progenitor cell subpopulations. Cell Stem Cell, 16(2):184–197. https://doi.org/10.1016/j.stem.2015.01.002
Patt S, Thiel G, Maas S, et al., 1993. Chromosomal changes and correspondingly altered proto-oncogene expression in human gliomas. Value of combined cytogenetic and molecular genetic analysis. Anticancer Res, 13(1):113–118.
Paz-Ares J, Ghosal D, Wienand U, et al., 1987. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J, 6(12):3553–3558. https://doi.org/10.1002/j.1460-2075.1987.tb02684.x
Ramsay RG, Gonda TJ, 2008. MYB function in normal and cancer cells. Nat Rev Cancer, 8(7):523–534. https://doi.org/10.1038/nrc2439
Rohs R, West SM, Sosinsky A, et al., 2009. The role of DNA shape in protein-DNA recognition. Nature, 461(7268):1248–1253. https://doi.org/10.1038/nature08473
Rosson D, Reddy EP, 1986. Nucleotide sequence of chicken c-myb complementary DNA and implications for myb oncogene activation. Nature, 319(6054):604–606.
Sarkar S, Singh PC, 2020. Alteration of the groove width of DNA induced by the multimodal hydrogen bonding of denaturants with DNA bases in its grooves affects their stability. Biochim Biophys Acta Gen Subj, 1864(3):129498. https://doi.org/10.1016/j.bbagen.2019.129498
Tan FE, Vladar EK, Ma L, et al., 2013. Myb promotes centriole amplification and later steps of the multiciliogenesis program. Development, 140(20):4277–4286. https://doi.org/10.1242/dev.094102
Tanikawa J, Yasukawa T, Enari M, et al., 1993. Recognition of specific DNA sequences by the c-myb protooncogene product: role of three repeat units in the DNA-binding domain. Proc Natl Acad Sci USA, 90(20):9320–9324. https://doi.org/10.1073/pnas.90.20.9320
Teleman O, Jönsson B, Engström S, 1987. A molecular dynamics simulation of a water model with intramolecular degrees of freedom. Mol Phys, 60(1):193–203. https://doi.org/10.1080/00268978700100141
Torelli G, Venturelli D, Coló A, et al., 1987. Expression of c-myb protooncogene and other cell cycle-related genes in normal and neoplastic human colonic mucosa. Cancer Res, 47(20):5266–5269.
Wallrapp C, Müller-Pillasch F, Solinas-Toldo S, et al., 1997. Characterization of a high copy number amplification at 6q24 in pancreatic cancer identifies c-myb as a candidate oncogene. Cancer Res, 57(15):3135–3139.
Wang S, Yang S, An BY, et al., 2011. Molecular dynamics analysis reveals structural insights into mechanism of nicotine N-demethylation catalyzed by tobacco cytochrome P450 mono-oxygenase. PLoS ONE, 6(8):e23342. https://doi.org/10.1371/journal.pone.0023342
Wolff L, 1996. Myb-induced transformation. Crit Rev Oncog, 7(3–4):245–260. https://doi.org/10.1615/critrevoncog.v7.i3-4.60
Xu LH, Zhao F, Yang WW, et al., 2019. MYB promotes the growth and metastasis of salivary adenoid cystic carcinoma. Int J Oncol, 54(5):1579–1590. https://doi.org/10.3892/ijo.2019.4754
Yang LW, Eyal E, Bahar I, et al., 2009. Principal component analysis of native ensembles of biomolecular structures (PCA_NEST): insights into functional dynamics. Bioinformatics, 25(5):606–614. https://doi.org/10.1093/bioinformatics/btp023
Zhao L, Glazov EA, Pattabiraman DR, et al., 2011. Integrated genome-wide chromatin occupancy and expression analyses identify key myeloid pro-differentiation transcription factors repressed by Myb. Nucleic Acids Res, 39(11):4664–4679. https://doi.org/10.1093/nar/gkr024
Acknowledgments
This work was supported by the National Key Research and Development Program of China (Nos. 2022YFC2402900 and 2022YFC2402901), the Fundamental Research Funds for the Central Universities (No. 226-2022-00213), and the Joint Funds of the Zhejiang Provincial Natural Science Foundation of China (No. LHDMD23H300001). We thank Tsukuba Life Sciences Center, the Institute of Physical and Chemical Research (RIKEN), Japan for the assistance on the quantitative EMSA experiments accomplished by Prof. Dongyun HAO during his post-doctoral study. We also thank Dr. Shichen WANG from the Texas A&M AgriLife Research (TX, USA) for the assistance on bioinformatics experiments.
Author information
Authors and Affiliations
Contributions
Shan WANG conceived, designed, and supervised the whole research, designed bioinformatics experiments, and wrote the manuscript with input from all authors. Dongyun HAO conceived and supervised the research, designed validation experiments, and wrote the manuscript. Jinru WENG carried out the bioinformatics analysis. Shuo YANG carried out the validation experiments. Jinkang SHEN, Hongsen LIU, and Yuzi XU participated in the bioinformatics analysis and interpretation of data. All authors have read and approved the final manuscript, and therefore, have full access to all the data in the study and take responsibility for the integrity and security of the data.
Corresponding authors
Ethics declarations
Shan WANG is a young scientist committee member for Journal of Zhejiang University-SCIENCE B (Biomedicine & Biotechnology) and was not involved in the editorial review or the decision to publish this article. Jinru WENG, Shuo YANG, Jinkang SHEN, Hongsen LIU, Yuzi XU, Dongyun HAO, and Shan WANG declare that they have no conflicts of interest.
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Supplementary information
Tables S1–S5; Figs. S1–S4
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Weng, J., Yang, S., Shen, J. et al. Molecular dynamics simulation reveals DNA-specific recognition mechanism via c-Myb in pseudo-palindromic consensus of mim-1 promoter. J. Zhejiang Univ. Sci. B 24, 883–895 (2023). https://doi.org/10.1631/jzus.B2200634
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B2200634
Key words
- c-Myb
- DNA-specific recognition mechanism
- Molecular dynamics simulation
- DNA major groove width
- Electrostatic potential