Abstract
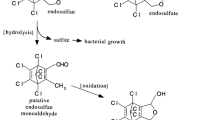
Atrazine is a selective herbicide used in agricultural fields to control the emergence of broadleaf and grassy weeds. The persistence of this herbicide is influenced by the metabolic action of habituated native microorganisms. This study provides information on the occurrence of atrazine mineralizing bacterial strains with faster metabolizing ability. The enrichment cultures were tested for the biodegradation of atrazine by high-performance liquid chromatography (HPLC) and mass spectrometry. Nine cultures JS01.Deg01 to JS09.Deg01 were identified as the degrader of atrazine in the enrichment culture. The three isolates JS04.Deg01, JS07.Deg01, and JS08.Deg01 were identified as efficient atrazine metabolizers. Isolates JS04.Deg01 and JS07.Deg01 produced hydroxyatrazine (HA) N-isopropylammelide and cyanuric acid by dealkylation reaction. The isolate JS08.Deg01 generated deethylatrazine (DEA), deisopropylatrazine (DIA), and cyanuric acid by N-dealkylation in the upper degradation pathway and later it incorporated cyanuric acid in their biomass by the lower degradation pathway. The optimum pH for degrading atrazine by JS08.Deg01 was 7.0 and 16S rDNA phylogenetic typing identified it as Enterobacter cloacae strain JS08.Deg01. The highest atrazine mineralization was observed in case of isolate JS08.Deg01, where an ample amount of trzD mRNA was quantified at 72 h of incubation with atrazine. Atrazine bioremediating isolate E. cloacae strain JS08.Deg01 could be the better environmental remediator of agricultural soils and the crop fields contaminated with atrazine could be the source of the efficient biodegrading microbial strains for the environmental cleanup process.
Similar content being viewed by others
References
Agertved, J., Rugge, K., Barker, J.F., 1992. Transformation of the herbicides MCPP and atrazine under natural aquifer conditions. Ground Water, 30(4):500–506. [doi:10.1111/j.1745-6584.1992.tb01525.x]
Behki, R.M., Khan, S.U., 1994. Degradation of atrazine, propazine, and simazine by Rhodococcus strain B-30. J. Agric. Food Chem., 42(5):1237–1241. [doi:10.1021/jf00041a036]
Behki, R., Topp, E., Dick, W., Germon, P., 1993. Metabolism of the herbicide atrazine by Rhodococcus strains. Appl. Environ. Microbiol., 59(6):1955–1959.
Cappuccino, J.G., Sherman, N., 2004. Microbiology: a Laboratory Manual. Pearson Education Inc., Singapore.
Cheng, G., Shapir, N., Sadowsky, M.J., Wackett, L.P., 2005. Allophanate hydrolase, not urease, functions in bacterial cyanuric acid metabolism. Appl. Environ. Microbiol., 71(8):4437–4445. [doi:10.1128/AEM.71.8.4437-4445.2005]
Cohen, S.Z., Creege, S.M., Carsel, R.F., Enfield, C.G., 1984. Potential Pesticide Contamination of Groundwater from Agricultural Uses. In: Krueger, R.F., Sieber, J.N. (Eds.), Treatment and Disposal of Pesticide Wastes. p.297–325. [doi:10.1021/bk-1984-0259.ch018]
Cole, J.R., Chai, B., Marsh, T.L., Farris, R.J., Wang, Q., Kulam, S.A., Chandra, S., McGarrell, D.M., Schmidt, T.M., Garrity, G.M., et al., 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucl. Acids Res., 31(1):442–443. [doi:10.1093/nar/gkg039]
Comber, S.D.W., 1999. Abiotic persistence of atrazine and simazine in water. Pestic. Sci., 55(7):696–702. [doi:10.1002/(SICI)1096-9063(199907)55:7〈696::AID-PS11〉3.0.CO;2-7]
Cook, A.M., 1987. Biodegradation of s-triazine xenobiotics. FEMS Microbiol. Lett., 46(2):93–116. [doi:10.1016/0378-1097(87)90059-0]
de Souza, M.L., Newcombe, D., Alvey, S., Crowley, D.E., Hay, A., Sadowsky, M.J., Wackett, L.P., 1998. Molecular basis of a bacterial consortium: interspecies catabolism of atrazine. Appl. Environ. Microbiol., 64(1):178–184.
Eaton, R.W., Karns, J.S., 1991. Cloning and analysis of the s-triazine catabolic genes from Pseudomonas sp. strain NRRLB-12227. J. Bacteriol., 173(3):1215–1222.
Felsenstein, J., 2008. PHYLIP (Phylogeny Inference Package), Version 3.68. Department of Genome Sciences, University of Washington, Seattle, USA.
Fruchey, I., Shapir, N., Sadowsky, M.J., Wackett, L.P., 2003. On the origins of cyanuric acid hydrolase: purification, substrates, and prevalence of AtzD from Pseudomonas sp. strain ADP. Appl. Environ. Microbiol., 69(6):3653–3657. [doi:10.1128/AEM.69.6.3653-3657.2003]
Ganesh-Kumar, S., Solomon, R.D.J., Kalimuthu, K., Vimalan, J., 2010. Evidence of aerobic degradation of polychlorinated biphenyl as growth substrate by new bacteria Stenotrophomonas sp. Jsg1 and Cupriavidus taiwanensis Jsg2. Carpath. J. Earth Environ. Sci., 5(2):169–176.
Ganesh-Kumar, S., Kalimuthu, K., Solomon, R.D.J., 2013. A novel bacterium that degrades Aroclor-1254 and its bphC gene encodes an extradiol aromatic ring cleavage dioxygenase (EARCD). Water Air Soil Pollut., 224(6):1587. [doi:10.1007/s11270-013-1587-0]
Ghosh, P.K., Philip, L.I.G.Y., 2006. Environmental significance of atrazine in aqueous systems and its removal by biological processes: an overview. Global NEST J., 8(2):159–178.
Hanioka, N., Jinno, H., Tanaka-Kagawa, T., Nishimura, T., Ando, M., 1999. In vitro metabolism of chlorotriazines: characterization of simazine, atrazine, and propazine metabolism using liver microsomes for rats treated with various cytochrome P450 inducers. Toxicol. Appl. Pharmacol., 156(3):195–205. [doi:10.1006/taap.1999.8648]
Hapeman, C.J., Karns, J.S., Shelton, D.R., 1995. Total mineralization of aqueous atrazine in the presence of ammonium nitrate using ozone and Klebsiella terragena (strain DRS-I): mechanistic considerations for pilot scale disposal. J. Agric. Food Chem., 43(5):1383–1391. [doi:10.1021/jf00053a047]
Kadian, N., Gupta, A., Satya, S., Mehta, R.K., Malik, A., 2008. Biodegradation of herbicide (atrazine) in contaminated soil using various bioprocessed materials. Bioresource Technol., 99(11):4642–4647. [doi:10.1016/j.biortech.2007.06.064]
Kalimuthu, K., Solomon, R.D.J., Ganesh-Kumar, S., Vimalan, J., 2011. Reductive dechlorination of perchloroethylene by Bacillus sp. Jsk1 isolated from dry cleaning industrial sludge. Carpath. J. Earth Environ. Sci., 6(1):165–170.
Karns, J.S., 1999. Gene sequence and properties of an s-triazine ring cleavage enzyme from Pseudomonas sp. strain NRRLB-12227. Appl. Environ. Microbiol., 65(8): 3512–3517.
Karns, J.S., Eaton, R.W., 1997. Genes encoding s-triazine degradation are plasmid-borne in Klebsiella pneumoniae strain 99. J. Agric. Food Chem., 45(3):1017–1022. [doi:10.1021/jf960464+]
Kolpin, D.W., Thurman, E.M., Linhart, S.M., 1998. The environmental occurrence of herbicides: the importance of degradates in groundwater. Arch. Environ. Contam. Toxicol., 35(3):385–390. [doi:10.1007/s002449900392]
Lone, M.I., He, Z.L., Stoffella, P.J., Yang, X.E., 2008. Phytoremediation of heavy metal polluted soils and water: progresses and perspectives. J. Zhejiang Univ.-Sci. B, 9(3):210–220. [doi:10.1631/jzus.B0710633]
Mandelbaum, R.T., Allan, D.L., Wackett, L.P., 1995. Isolation and characterization of a Pseudomonas sp. that mineralizes the s-triazine herbicide atrazine. Appl. Environ. Microbiol., 61(4):1451–1457.
Martinez, B., Tomkins, J., Wackett, L.P., Wing, R., Sadowsky, M.J., 2001. Complete nucleotide sequence and organization of the atrazine catabolic plasmid pADP-1 from Pseudomonas sp. strain ADP. J. Bacteriol., 83(19): 5684–5697. [doi:10.1128/JB.183.19.5684-5697.2001]
McInroy, J.A., Kloepper, J.W., 1995. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil, 173(2):337–342. [doi:10.1007/BF00011472]
Piutti, S., Semon, E., Landry, D., Hartmann, A., Dousset, S., Lichtfouse, E., Topp, E., Soulas, G., Martin-Laurent, F., 2003. Isolation and characterisation of Nocardioides sp. SP12, an atrazine-degrading bacterial strain possessing the gene trzN from bulk and maize rhizosphere soil. FEMS Microbiol. Lett., 221(1):111–117. [doi:10.1016/S0378-1097(03)00168-X]
Popov, V.H., Cornish, P.S., Sultana, K., Morris, E.C., 2005. Atrazine degradation in soils: the role of microbial communities, atrazine application history, and soil carbon. Soil Res., 43(7):861–871. [doi:10.1071/SR04048]
Rousseaux, S., Hartmann, A., Soulas, G., 2001. Isolation and characterisation of new Gram-negative and Gram- positive atrazine degrading bacteria from different French soils. FEMS Microbiol. Ecol., 36(2–3):211–222. [doi:10.1016/S0168-6496(01)00135-0]
Sambrook, J., Russel, D.W., 2001. Molecular Cloning: a Laboratory Manual, 3rd Ed. Cold Spring Harbor Laboratory Press, p.213.
Satheeja Santhi, V., Jebakumar, S.R.D., 2011. Phylogenetic analysis and antimicrobial activities of Streptomyces isolates from mangrove sediment. J. Basic Microbiol., 51(1): 71–79. [doi:10.1002/jobm.201000107]
Seghers, D., Wittebolle, L., Top, E.M., Verstraete, W., Siciliano, S.D., 2004. Impact of agricultural practices on the Zea mays L. endophytic community. Appl. Environ. Microbiol., 70(3):1475–1482. [doi:10.1128/AEM.70.3.1475-1482.2004]
Seiler, A., Brenneisen, P., Green, D.H., 1992. Benefits and risks of plant protection products-possibilities of protecting drinking water: case atrazine. Water Supply, 10(2):31–42.
Seto, M., Kimbara, K., Shimura, M., Hatta, T., Fukuda, M., Yano, K., 1995. A novel transformation of polychlorinated biphenyls by Rhodococcus sp. strain RHA1. Appl. Environ. Microbiol., 61(9):3353–3358.
Shapir, N., Osborne, J.P., Johnson, G., Sadowsky, M.J., Wackett, L.P., 2002. Purification, substrate range, and metal center of AtzC: the N-isopropylammelide aminohydrolase involved in bacterial atrazine metabolism. J. Bacteriol., 184(19):5376–5384. [doi:10.1128/JB.184.19.5376-5384.2002]
Shapir, N., Cheng, G., Sadowsky, M.J., Wackett, L.P., 2006. Purification and characterization of TrzF: biuret hydrolysis by allophanate hydrolase supports growth. Appl. Environ. Microbiol., 72(4):2491–2495. [doi:10.1128/AEM. 72.4.2491-2495.2006]
Siripattanakul, S., Wirojanagud, W., McEvoy, J., Limpiyakorn, T., Khan, E., 2009. Atrazine degradation by stable mixed cultures enriched from agricultural soil and their characterization. J. Appl. Microbiol., 106(3):986–992. [doi:10. 1111/j.1365-2672.2008.04075.x]
Stackebrandt, E., Goebel, M., 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Bacteriol., 44(4):846–849. [doi:10.1099/00207713-44-4-846]
Struthers, J.K., Jayachandran, K., Moorman, T.B., 1998. Biodegradation of atrazine by Agrobacterium radiobacter J14a and use of this strain in bioremediation of contaminated soil Appl. Environ. Microbiol., 64(9):3368–3375.
Summers, W., 1970. A simple method for extraction of RNA from E. coli utilizing diethylpyrocarbonate. Anal. Biochem., 33(2):459–463.
Suzuki, D., Baba, D., Satheeja Santhi, V., Solomon, R.D.J., Katayama, A., 2013. Use of a glass bead-containing liquid medium for efficient production of a soil-free culture with polychlorinated biphenyl-dechlorination activity. World J. Microbiol. Biotechnol., 29(8):1461–1471. [doi:10.1007/s11274-013-1310-8]
Topp, E., Zhu, H., Nour, S.M., Houot, S., Lewis, M., Cuppels, D., 2000. Characterization of an atrazine-degrading Pseudaminobacter sp. isolated from Canadian and French agricultural soils. Appl. Environ. Microbiol., 66(7): 2773–2782. [doi:10.1128/AEM.66.7.2773-2782.2000]
Udiković-Kolić, N., Hršak, D., Devers, M., Klepac-Ceraj, V., Petrić, I., Martin-Laurent, F., 2010. Taxonomic and functional diversity of atrazine-degrading bacterial communities enriched from agrochemical factory soil. J. Appl. Microbiol., 109(1):355–367. [doi:10.1111/j.1365-2672.2010.04700.x]
van Zwieten, L., Kennedy, I.R., 1995. Rapid degradation of atrazine by Rhodococcus sp. NI86/21 and by an atrazine-perfused soil. J. Agric. Food Chem., 43(5):1377–1382. [doi:10.1021/jf00053a046]
Wackett, L., Sadowsky, M., Martinez, B., Shapir, N., 2002. Biodegradation of atrazine and related s-triazine compounds: from enzymes to field studies. Appl. Microbiol. Biotechnol., 58(1):39–45. [doi:10.1007/s00253-001-0862-y]
Wang, J.H., Zhu, L.S., Liu, A.J., Ma, T.T., Wang, Q., Xie, H., Wang, J., Jiang, T., Zhao, R.S., 2011. Isolation and characterization of an Arthrobacter sp. strain HB-5 that transforms atrazine. Environ. Geochem. Health, 33(3): 259–266. [doi:10.1007/s10653-010-9337-3]
Wang, Q., Xie, S., 2012. Isolation and characterization of a high-efficiency soil atrazine-degrading Arthrobacter sp. strain. Int. Biodeter. Biodegr., 71:61–66. [doi:10.1016/j.ibiod.2012.04.005]
Widmer, S.K., Spalding, R.F., 1995. A natural gradient transport study of selected herbicides. J. Environ. Qual., 24(3): 445–453.
Yanze-Kontchou, C., Gschwind, N., 1994. Mineralization of the herbicide atrazine as a carbon source by a Pseudomonas strain. Appl. Environ. Microbiol., 60(12): 4297–4302.
Zhang, S.Y., Wang, Q.F., Wan, R., Xie, S.G., 2011. Changes in bacterial community of anthracene bioremediation in municipal solid waste composting soil. J. Zhejiang Univ.-Sci. B (Biomed. & Biotechnol.), 12(9):760–768. [doi:10.1631/jzus.B1000440]
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Solomon, R.D.J., Kumar, A. & Satheeja Santhi, V. Atrazine biodegradation efficiency, metabolite detection, and trzD gene expression by enrichment bacterial cultures from agricultural soil. J. Zhejiang Univ. Sci. B 14, 1162–1172 (2013). https://doi.org/10.1631/jzus.B1300001
Received:
Revised:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1300001