Abstract
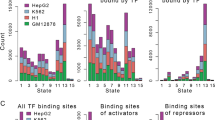
OCT4, a member of the POU family of gene products, is an octamer motif-binding transcription factor. As it is known to play a crucial role in cancer processes including proliferation, invasion, and chemoradioresistance, it is important to identify the direct targets of OCT4 in living cancer cells. Here, chromatin immunoprecipitation-sequencing (ChIP-seq) was used to identify OCT4 binding sites in glioblastoma cancer cells. The results showed that 5438 OCT4 binding sites were localized in the glioblastoma cancer genome and that these sites contained a consensus sequence TTTkswTw (k=T or G, s=C or G, w=A or T), which occurred 3931 times in 2312 OCT4 binding regions. Furthermore, binding motifs of some other transcription factors were identified in OCT4 binding regions. Our results provide a valuable dataset for understanding gene regulation mechanisms underlying the function of OCT4 in glioblastoma cancer.
Similar content being viewed by others
References
Atlasi, Y., Mowla, S.J., Ziaee, S.A., Bahrami, A.R., 2007. OCT-4, an embryonic stem cell marker, is highly expressed in bladder cancer. Int. J. Cancer, 120(7): 1598–1602. [doi:10.1002/ijc.22508]
Barski, A., Zhao, K., 2009. Genomic location analysis by ChIP-seq. J. Cell. Biochem., 107(1):11–18. [doi:10.1002/jcb.22077]
Barski, A., Cuddapah, S., Cui, K., Roh, T.Y., Schones, D.E., Wang, Z., Wei, G., Chepelev, I., Zhao, K., 2007. High-resolution profiling of histone methylations in the human genome. Cell, 129(4):823–837. [doi:10.1016/j.cell.2007.05.009]
Boyer, L.A., Lee, T.I., Cole, M.F., Johnstone, S.E., Levine, S.S., Zucker, J.P., Guenther, M.G., Kumar, R.M., Murray, H.L., Jenner, R.G., et al., 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell, 122(6):947–956. [doi:10.1016/j.cell.2005.08.020]
Chen, Y.C., Hsu, H.S., Chen, Y.W., Tsai, T.H., How, C.K., Wang, C.Y., Hung, S.C., Chang, Y.L., Tsai, M.L., Lee, Y.Y., et al., 2008. Oct-4 expression maintained cancer stem-like properties in lung cancer-derived CD133-positive cells. PLoS One, 3(7):e2637. [doi:10.1371/journal.pone.0002637]
Cheng, C.J., Wu, Y.C., Shu, J.A., Ling, T.Y., Kuo, H.C., Wu, J.Y., Chang, E.E., Chang, S.C., Huang, Y.H., 2007. Aberrant expression and distribution of the OCT-4 transcription factor in seminomas. J. Biomed. Sci., 14(6):797–807. [doi:10.1007/s11373-007-9198-7]
Crooks, G.E., Hon, G., Chandonia, J.M., Brenner, S.E., 2004. WebLogo: a sequence logo generator. Genome Res., 14(6):1188–1190. [doi:10.1101/gr.849004]
Dekker, N., Cox, M., Boelens, R., Verrijzer, C.P., van der Vliet, P.C., Kaptein, R., 1993. Solution structure of the POU-specific DNA-binding domain of Oct-1. Nature, 362(6423):852–855. [doi:10.1038/362852a0]
Fang, X., Yu, W., Li, L., Shao, J., Zhao, N., Chen, Q., Ye, Z., Lin, S.C., Zheng, S., Lin, B., 2010. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. OMICS, 14(4):369–384. [doi:10.1089/omi.2010.0053]
Fang, X., Yoon, J., Li, L., Yu, W., Shao, J., Hua, D., Zheng, S., Hood, L., Goodlett, D.R., Foltz, G., Lin, B., 2011. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics, 12(1):11. [doi:10.1186/1471-2164-12-11]
Gilchrist, D.A., Fargo, D.C., Adelman, K., 2009. Using ChIP-chip and ChIP-seq to study the regulation of gene expression: genome-wide localization studies reveal widespread regulation of transcription elongation. Methods, 48(4):398–408. [doi:10.1016/j.ymeth.2009.02.024]
Herr, W., Cleary, M.A., 1995. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev., 9(14):1679–1693. [doi:10.1101/gad.9.14.1679]
Ji, H., Jiang, H., Ma, W., Johnson, D.S., Myers, R.M., Wong, W.H., 2008. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol., 26(11): 1293–1300. [doi:10.1038/nbt.1505]
Johnson, D.S., Mortazavi, A., Myers, R.M., Wold, B., 2007. Genome-wide mapping of in vivo protein-DNA interactions. Science, 316(5830):1497–1502. [doi:10.1126/science.1141319]
Kim, J.B., Greber, B., Arauzo-Bravo, M.J., Meyer, J., Park, K.I., Zaehres, H., Scholer, H.R., 2009a. Direct reprogramming of human neural stem cells by OCT4. Nature, 461(7264):643–649. [doi:10.1038/nature08436]
Kim, J.B., Sebastiano, V., Wu, G., Arauzo-Bravo, M.J., Sasse, P., Gentile, L., Ko, K., Ruau, D., Ehrich, M., van den Boom, D., et al., 2009b. Oct4-induced pluripotency in adult neural stem cells. Cell, 136(3):411–419. [doi:10.1016/j.cell.2009.01.023]
Kraft, H.J., Mosselman, S., Smits, H.A., Hohenstein, P., Piek, E., Chen, Q., Artzt, K., van Zoelen, E.J., 1996. Oct-4 regulates alternative platelet-derived growth factor α receptor gene promoter in human embryonal carcinoma cells. J. Biol. Chem., 271(22):12873–12878. [doi:10.1074/jbc.271.22.12873]
Lin, B., Wang, J., Hong, X., Yan, X., Hwang, D., Cho, J.H., Yi, D., Utleg, A.G., Fang, X., Schones, D.E., et al., 2009. Integrated expression profiling and ChIP-seq analyses of the growth inhibition response program of the androgen receptor. PLoS One, 4(8):e6589. [doi:10.1371/journal. pone.0006589]
Marson, A., Levine, S.S., Cole, M.F., Frampton, G.M., Brambrink, T., Johnstone, S., Guenther, M.G., Johnston, W.K., Wernig, M., Newman, J., et al., 2008. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell, 134(3):521–533. [doi:10.1016/j.cell.2008.07.020]
Monk, M., Holding, C., 2001. Human embryonic genes re-expressed in cancer cells. Oncogene, 20(56):8085–8091. [doi:10.1038/sj.onc.1205088]
Okumura-Nakanishi, S., Saito, M., Niwa, H., Ishikawa, F., 2005. Oct-3/4 and Sox2 regulate Oct-3/4 gene in embryonic stem cells. J. Biol. Chem., 280(7):5307–5317. [doi:10.1074/jbc.M410015200]
Pesce, M., Wang, X., Wolgemuth, D.J., Scholer, H., 1998. Differential expression of the Oct-4 transcription factor during mouse germ cell differentiation. Mech. Dev., 71(1–2):89–98. [doi:10.1016/S0925-4773(98)00002-1]
Schoorlemmer, J., Jonk, L., Sanbing, S., van Puijenbroek, A., Feijen, A., Kruijer, W., 1995. Regulation of Oct-4 gene expression during differentiation of EC cells. Mol. Biol. Rep., 21(3):129–140. [doi:10.1007/BF00997235]
Schreiber, E., Merchant, R.E., Wiestler, O.D., Fontana, A., 1994. Primary brain tumors differ in their expression of octamer deoxyribonucleic acid-binding transcription factors from long-term cultured glioma cell lines. Neurosurgery, 34(1):129–135. [doi:10.1227/00006123-199401000-00019]
Takahashi, K., Yamanaka, S., 2006. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell, 126(4):663–676. [doi:10.1016/j.cell.2006.07.024]
Takahashi, K., Tanabe, K., Ohnuki, M., Narita, M., Ichisaka, T., Tomoda, K., Yamanaka, S., 2007. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell, 131(5):861–872. [doi:10.1016/j.cell.2007.11.019]
Tay, Y., Zhang, J., Thomson, A.M., Lim, B., Rigoutsos, I., 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature, 455(7216):1124–1128. [doi:10.1038/nature07299]
Verrijzer, C.P., Alkema, M.J., van Weperen, W.W., van Leeuwen, H.C., Strating, M.J., van der Vliet, P.C., 1992. The DNA binding specificity of the bipartite POU domain and its subdomains. EMBO J., 11(13):4993–5003.
Visel, A., Blow, M.J., Li, Z., Zhang, T., Akiyama, J.A., Holt, A., Plajzer-Frick, I., Shoukry, M., Wright, C., Chen, F., et al., 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature, 457(7231):854–858. [doi: 10.1038/nature07730]
Wang, W., Zhang, P., Liu, X., 2009. Short read DNA fragment anchoring algorithm. BMC Bioinformatics, 10(s1):S17. [doi:10.1186/1471-2105-10-S1-S17]
Williams, D.C.Jr., Cai, M., Clore, G.M., 2004. Molecular basis for synergistic transcriptional activation by Oct1 and Sox2 revealed from the solution structure of the 42-kDa Oct1.Sox2.Hoxb1-DNA ternary transcription factor complex. J. Biol. Chem., 279(2):1449–1457. [doi:10.1074/jbc.M309790200]
Yu, J., Vodyanik, M.A., Smuga-Otto, K., Antosiewicz-Bourget, J., Frane, J.L., Tian, S., Nie, J., Jonsdottir, G.A., Ruotti, V., Stewart, R., et al., 2007. Induced pluripotent stem cell lines derived from human somatic cells. Science, 318(5858):1917–1920. [doi:10.1126/science.1151526]
Zeeberg, B.R., Qin, H., Narasimhan, S., Sunshine, M., Cao, H., Kane, D.W., Reimers, M., Stephens, R.M., Bryant, D., Burt, S.K., et al., 2005. High-throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID). BMC Bioinformatics, 6(1):168. [doi:10.1186/1471-2105-6-168]
Author information
Authors and Affiliations
Corresponding authors
Additional information
The two authors contributed equally to this work
Project supported by the Ministry of Science and Technology, China (Nos. 2004CB518707, 2006DFA32950, 2006AA02Z4A2, 2006AA02 A303, 2007DFC30360, and 2008DFA11320), and the National Natural Science Foundation of China (No. 81101580)
Electronic supplementary materials: The online version of this article (doi:10.1631/jzus.B1100059) contains supplementary materials, which are available to authorized users
Rights and permissions
About this article
Cite this article
Fang, Xf., Zhang, Wy., Zhao, N. et al. Genome-wide analysis of OCT4 binding sites in glioblastoma cancer cells. J. Zhejiang Univ. Sci. B 12, 812–819 (2011). https://doi.org/10.1631/jzus.B1100059
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1631/jzus.B1100059