Abstract
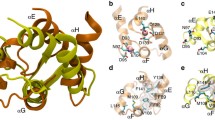
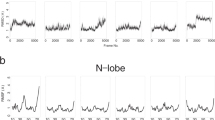
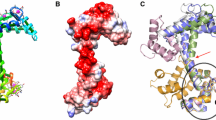
We analyze the apo and holo calmodulin (CaM) structures by sequentially inserting a perturbation on every residue of the protein, and monitoring the linear response. Residue crosscorrelation matrices obtained from 20 ns long molecular dynamics simulation of the apo-form are used as the kernel in the linear response. We determine two residues whose perturbation equivalently yields the experimentally determined displacement profiles of CaM, relevant to the binding of the trifluoperazine (TFP) ligand. They reside on structurally equivalent positions on the N- and C-terminus lobes of CaM, and are not in direct contact with the binding region. The direction of the perturbation that must be inserted on these residues is an important factor in recovering the conformational change, implying that highly selective binding must occur near these sites to invoke the necessary conformational change.
Similar content being viewed by others
References
M. Ikura, J.B. Ames, Proc. Natl. Acad. Sci. 103,1159–1164 (2006).
J.L. Fallon, F.A. Quicho, Structure 11, 1303–1307 (2003).
M. Vandonselaar, R.A. Hickie, W.J. Quail and T.J. Delbaere, Struct. Biol. 1, 795–801 (1994).
CM. Shepherd and H.J. Vogel, Blophys. J. 87, 780–791 (2004).
C. Atilgan, A.R. Atilgan, PLoS Comput. Biol. 5, el000544 (2009).
M. Ikeguchi, J. Ueno, M. Sato, and A. Kidera, Phys. Rev. Lett. 94, 078102 (2005).
L.S. Yilmaz, A.R. Atilgan, J. Chem. Phys. 113, 4454–4464 (2000).
C. Atilgan, Z.N. Gerek, S.B. Ozkan, A.R. Atilgan, Blophys. J. 99, 933–943 (2010).
A.R. Atilgan, S.R. Durell, R.L. Jernigan, M.C. Demirei, O. Keskin, et al, Blophys. J. 80, 505–515(2001).
C. Baysal, A.R. Atilgan, Proteins. 45, 62–70 (2001).
CBaysal, A.R. Atilgan, Proteins. 43, 150–160 (2001).
Y.S. Babu, C.E. Bugg , W.J. Cook, J. Mol. Biol. 204, 191–204 (1988).
W. Humphrey, A. Dalke, K. Schuiten, J. Mol. Graph 14, 33–38(1996).
J.C Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, et al., J. Comput. Chem. 26, 1781–1802(2005).
B.R. Brooks, R.E. Bruccoleri, B.D. Olafson, D.J. States, S. Swaminathan, et al., J. Comput. Chem. 4, 187–217 (1983).
T. Darden, L. Perera, L.P. Li, L. Pedersen, Struc. Fold.Des. 7, R55–R60 (1999).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Aykut, A.O., Atilgan, A.R. & Atilgan, C. Molecular Recognition Mechanisms of Calmodulin Examined by Perturbation-Response Scanning. MRS Online Proceedings Library 1301, 137–142 (2011). https://doi.org/10.1557/opl.2011.568
Published:
Issue Date:
DOI: https://doi.org/10.1557/opl.2011.568