Abstract
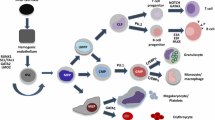
Hematopoiesis has provided a valuable model for examining how genetic programs are established and executed in terms of cell fate decision. Identification of common myeloid and lymphoid progenitors allows us to directly assess the regulatory mechanisms of lineage commitment. Multiple markers of hematopoietic lineages are coexpressed in hematopoietic stem cells and progenitors, a phenomenon referred to as lineage priming. Promiscuous expression of several lineage-affiliated genes precedes lineage commitment but does not alter the biological potential of hematopoietic stem cells and multipotent progenitors. Promiscuous accessibility of multiple programs allows flexibility in cell fate commitment at the multipotent stages, indicating that transcriptional promiscuity can operate in stem cells and progenitors to control their transition from multipotency to single-line age commitment.
Similar content being viewed by others
References
Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168.
Morrison SJ, Weissman IL. The long-term repopulating subset of hematopoietic stem cells is deterministic and isolatable by pheno- type. Immunity. 1994;l:661–673.
Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(-)Scal(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15:659–669.
Christensen JL, Weissman IL. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc Natl Acad Sci U S A. 2001;98:14541–14546.
Metcalf D. Stem cells, pre-progenitor cells and lineage-committed cells: are our dogmas correct? Ann N Y Acad Sci. 1999;872:289–303.
Orkin SH. Diversification of haematopoietic stem cells to specific lineages. Nat Rev Genet. 2000;l:57–64.
Akashi K, He X, Chen J, et al. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389.
Terskikh AV, Miyamoto T, Chang C, Diatchenko L, Weissman IL. Gene expression analysis of purified hematopoietic stem cells and committed progenitors. Blood. 2003;101:383–389.
Hu M, Krause D, Greaves M, et al. Multilineage gene expression precedes commitment in the hemopoietic system. Genes Dev. 1997; 11:774–785.
Enver T, Greaves M. Loops, lineage, and leukemia. Cell. 1998;94:9–12.
Enver T, Heyworth CM, Dexter TM. Do stem cells play dice? Blood. 1998;92:348–351.
Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997; 91:661–672.
Akashi K, Traver D, Miyamoto T, Weissman IL. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature. 2000;404:193–197.
Noguchi M, Nakamura Y, Russell SM, et al. Interleukin-2 receptor gamma chain: a functional component of the interleukin-7 receptor. Science. 1993;262:1877–1880.
Peschon JJ, Morrissey PJ, Grabstein KH, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mics. J Exp Med. 1994;180:1955–1960.
von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Murray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526.
Akashi K, Kondo M, von Freeden-Jeffry U, Murray R, Weissman IL. Bcl-2 rescues T lymphopoiesis in interleukin-7 receptor-deficient mice. Cell. 1997;89:1033–1041.
Akashi K, Kondo M, Weissman IL. Role of interleukin-7 in T-cell development from hematopoietic stem cells. Immunol Rev. 1998; 165:13–28.
Corcoran AE, Riddell A, Krooshoop D, Venkitaraman AR. Impaired immunoglobulin gene rearrangement in mice lacking the IL-7 receptor. Nature. 1998;391:904–907.
Felsenfeld G, Boyes J, Chung J, Clark D, Studitsky V. Chromatin structure and gene expression. Proc Natl Acad Sci U S A. 1996;93:9384–9388.
Berger SL, Felsenfeld G. Chromatin goes global. Mol Cell. 2001;8:263–268.
Weintraub H. Assembly and propagation of repressed and depressed chromosomal states. Cell. 1985;42:705–711.
Kontaraki J, Chen HH, Riggs A, Bonifer C. Chromatin fine structure profiles for a developmentally regulated gene: reorganization of the lysozyme locus before trans-activator binding and gene expression. Genes Dev. 2000;14:2106–2122.
Cross MA, Enver T. The lineage commitment of haemopoietic pro- genitor cells. Curr Opin Genet Dev. 1997;7:609–613.
Jimenez G, Griffiths SD, Ford AM, Greaves MF, Enver T. Activation of the beta-globin locus control region precedes commitment to the erythroid lineage. Proc Natl Acad Sci U S A. 1992;89:10618–10622.
Delassus S, Titley I, Enver T. Functional and molecular analysis of hematopoietic progenitors derived from the aorta-gonad- mesonephros region of the mouse embryo. Blood. 1999;94:1495–1503.
Shivdasani RA, Fujiwara Y, McDevitt MA, Orkin SH. A lineage- selective knockout establishes the critical role of transcription factor GATA-1 in megakaryocyte growth and platelet development. EMBO J. 1997;16:3965–3973.
Yamaguchi Y, Zon LI, Ackerman SJ, Yamamoto M, Suda T. Forced GATA-1 expression in the murine myeloid cell line Ml: induction of c-Mpl expression and megakaryocytic/erythroid differentiation. Blood. 1998;91:450–457.
Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 1994;371:221–226.
Shivdasani RA, Rosenblatt MF, Zucker-Franklin D, et al. Transcription factor NF-E2 is required for platelet formation independent of the actions of thrombopoietin/MGDF in megakaryocyte development. Cell. 1995;81:695–704.
Zhang DE, Zhang P, Wang ND, Hetherington CJ, Darlington GJ, Tenen DG. Absence of granulocyte colony-stimulating factor signaling and neutrophil development in CCAAT enhancer binding protein alpha-deficient mice. Proc Natl Acad Sci U S A. 1997;94:569–574.
Tenen DG, Hromas R, Licht JD, Zhang DE. Transcription factors, normal myeloid development, and leukemia. Blood. 1997;90:569–574.
Morgan B, Sun L, Avitahl N, et al. Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J. 1997;16:2004–2013.
Ho IC, Vorhees P, Marin N, et al. Human GATA-3: a lineage- restricted transcription factor that regulates the expression of the T cell receptor alpha gene. EMBO J. 1991;10:1187–1192.
Nutt SL, Heavey B, Rolink AG, Busslinger M. Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature. 1999;401:556–562.
Allen RD, 3rd, Bender TP, Siu G. c-Myb is essential for early T cell development. Genes Dev. 1999;13:1073–1078.
Badiani P, Corbella P, Kioussis D, Marvel J, Weston K. Dominant interfering alleles define a role for c-Myb in T-cell development. Genes Dev. 1994;8:770–782.
Miyamoto T, Iwasaki H, Reizis B, et al. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Dev Cell. 2002;3:137–147.
van den Elsen P, Bruns G, Gerhard DS, et al. Assignment of the gene coding for the T3-delta subunit of the T3-T-cell receptor complex to the long arm of human chromosome 11 and to mouse chromosome 9. Proc Natl Acad Sci U S A. 1985;82:2920–2924.
Sakaguchi N, Melchers F. Lambda 5, a new light-chain-related locus selectively expressed in pre-B lymphocytes. Nature. 1986;324:579–582.
Karasuyama H, Rolink A, Shinkai Y, Young F, Alt FW, Melchers F. The expression of Vpre-B/lambda 5 surrogate light chain in early bone marrow precursor B cells of normal and B cell-deficient mutant mice. Cell. 1994;77:133–143.
Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726.
Ye M, Iwasaki H, Laiosa CV, et al. Hematopoietic stem cells expressing the myeloid lysozyme gene retain long-term, multilin- eage repopulation potential. Immunity 2003;19:689–699.
Gounari F, Aifantis I, Martin C, et al. Tracing lymphopoiesis with the aid of a pTalpha-controlled reporter gene. Nat Immunol. 2002; 3:489–496.
Iwasaki H, Mizuno S, Wells RA, Cantor AB, Watanabe S, Akashi K. GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity. 2003;19:451–462.
Nerlov C, Graf T. PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 1998;12:2403–2412.
Zhang P, Behre G, Pan J, et al. Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc Natl Acad Sci U S A. 1999;96:8705–8710.
Zhang P, Zhang X, Iwama A, et al. PU.l inhibits GATA-1 function and erythroid differentiation by blocking GATA-1 DNA binding. Blood. 2000;96:2641–2648.
Kulessa H, Frampton J, Graf T. GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 1995;9:1250–1262.
DeKoter RP, Singh H. Regulation of B lymphocyte and macrophage development by graded expression of PU.l. Science. 2000; 288:1439–1441.
Rothenberg EV, Dionne CJ. Lineage plasticity and commitment in T-cell development. Immunol Rev. 2002;187:96–115.
Wagers AJ, Weissman IL. Plasticity of adult stem cells. Cell. 2004; 116:639–648.
Kondo M, Scherer DC, Miyamoto T, et al. Cell-fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature. 2000;407:383–386.
King AG, Kondo M, Scherer DC, Weissman IL. Lineage infidelity in myeloid cells with TCR gene rearrangement: a latent developmental potential of proT cells revealed by ectopic cytokine receptor signaling. Proc Natl Acad Sci U S A. 2002;99:4508–4513.
Iwasaki-Arai J, Iwasaki H, Miyamoto T, Watanabe S, Akashi K. Enforced granulocyte/macrophage colony-stimulating factor signals do not support lymphopoiesis, but instruct lymphoid to myelomonocytic lineage conversion. J Exp Med. 2003;197:1311–1322.
Author information
Authors and Affiliations
Corresponding author
About this article
Cite this article
Miyamoto, T., Akashi, K. Lineage Promiscuous Expression of Transcription Factors in Normal Hematopoiesis. Int J Hematol 81, 361–367 (2005). https://doi.org/10.1532/IJH97.05003
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1532/IJH97.05003