Abstract
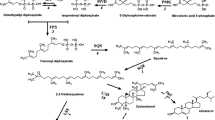
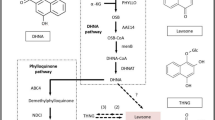
Indigo is the most important blue dye for textile dyeing and is biosynthesized in Polygonum tinctorium. Some biochemical studies related to biosynthesis are available. However, genomic and transcriptome studies have not received sufficient attention. Here, we report de novo assembly of transcriptome datasets and its comprehensive analysis. A total of 60,395 unigenes were annotated using BLAST search against the different databases. At least 23,721 unigenes mapped onto different pathways using KEGG database. We found that 3,323 genes are involved in biosynthesis of secondary metabolites, 117 phenylalanine, tyrosine and tryptophan biosynthesis and 147 tryptophan metabolisms. Apart from this, indigo biosynthesis pathway genes viz., dioxygenase, monooxygenase, and glucosyltransferase have also been identified. Fourteen genes encoding cytochrome P450 monooxygenase, 26 glucoside dioxygenase, 9 UDP-glucose D-glucosyltransferase and 52 were β-D-glucosidase. These findings provide a foundation for further analysis of this pathway with potential to enhance the synthesis of indican in P. tinctorium.
Similar content being viewed by others
Abbreviations
- COG:
-
clusters of orthologous group
- GO:
-
gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- NR:
-
non-redundant protein database
- NT:
-
non-redundant nucleotide database
- UDP:
-
uridine diphosphate glucose
References
Conesa A., Gotz S., GarrciaGomez M., Terol J., Talon M. & Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676.
Ensley B.D., Ratzkin B.J., Osslund T.D., Simon M.J., Wackett L.P. & Gibson D.T. 1983. Expression of naphthalene oxidation genes in Escherichio, coli results in the biosynthesis of indigo. Science 222: 167–169.
Epstein E., Nabors M.W. & Stowe B.B. 1967. Origin of indigo of woad. Nature 216: 547–549.
Garg R., Patel R.K., Tyagi A.K. & Jain M. 2011. De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res. 18: 53–63.
Grabherr M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., Chen Z., Mauceli E., Hacohen N., Gnirke A., Rhind N., Palma F., Birren B.W., Nusbaum C., LindbladToh K., Friedman N. & Regev A. 2011. Fulllength transcriptome assembly from RNASeq data without a reference genome. Nature Biotechnol. 29: 644–652.
Kanehisa M., Araki M., Goto S., Hattori M., Hirakawa M., Itoh M., Katayama T., Kawashima S., Okuda S., Tokimatsu T. & Yamanishi Y. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 36: D480-D484.
Kim J.Y., Lee J.Y., Shin Y. & Kim G.J. 2010. Characterization of an indicanthydrolyzing enzyme from Sinorhizobium, meliloti. Process Biochem. 45: 892–896.
Kudapa H., Bharti A.K., Cannon S.B., Farmer A.D., Mulaosmanovic B., Kramer R., Bohra A., Weeks N.T., Crow J.A., Tuteja R., Shah T., Dutta S., Gupta D.K., Singh A., Gaikwad K., Sharma T.R., May G.D., Singh N.K. & Varshney R.K. 2012. A comprehensive transcriptome assembly of pigeonpea (Cajanus cajan L.) using Sanger and secondgeneration sequencing platforms. Mol. Plant. 5: 1020–1028.
Lee C.Y. & Kim W.J. 1990. Production of natural colorants by plant cell biotechnology, pp. 81-85. In: Natural Spices and Pigments (in Korean), Hyangmoon Publishing Co., Seoul, Korea.
Marcinek H., Weyler W., DeusNeumann B. & Zenk M.H. 2000. IndoxylUDPGglucosyltransferase from Baphicacanthus cusia. Phytochemistry 53: 201–207.
Maugard T., Enaud E., Sayette A.L., Choisy P. & Legoy M.D. 2002. βGlucosidase catalysed hydrolysis of indican from leaves of Polygonum tinctorium. Biotechnol. Progr. 18: 1104–1108.
Minami Y., Kanafuji T. & Miura K. 1996. Puri fication and characterization of a βglucosidase from Polygonum, tinctorium, which catalyzes preferentially the hydrolysis of indican. Biosci. Biotech. Biochem. 60: 147–149.
Minami Y., Nishimura O., HaraNishimura I., Nishimura M. & Matsubara H. 2000. Tissue and intracellular localization of indican and the puri fication and characterization of indican synthase from indigo plants. Plant Cell Physiol. 41: 218–225.
Minami Y., Shigeta Y., Tokumoto U., Tanaka Y., YonekuraSakakibara K. & Ohoka H. 1999. Cloning, sequencing, characterization, and expression of a βglucosidase cDNA from the indigo plant. Plant Sci. 142: 219–226.
Minami Y., Takao H., Kanafuji T., Miura K., Kondo M., HaraNishimura I., Nishimura M. & Matsubara, H. 1997. βGlucosidase in the indigo plant: intracellular localization and tissue speci fic expression in leaves. Plant Cell Physiol. 38: 1069–1074.
Peng H. & Zhang J. 2009. Commercial highthroughput sequencing and its applications in DNA analysis. Biologia 64: 20–26.
Schullehner K., Dick R., Vitzthum F., Schwab W., Brandt W., Frey M. & Gierl A. 2008. Benzoxazinoid biosynthesis in dicot plants. Phytochemistry 69: 2668–2677.
Shin Y., Yoo D.I. & Kim K. 2012. Process balance of natural indigo production based on traditional Niram method. Textile Coloration and Finishing 24: 253–259.
Song K.S., Shim J.Y., Jung D.S. & Kim S.U. 2011. Origin of oxygen in indoxylderivatives of Polygonum tinctorium L. as probed by 18O2 feeding. J. Korean Soc. Appl. Biol. Chem. 54: 340–344.
Stoker K.G., Cooke D.T. & Hill D.J. 1998. An improved method for the largescale processing of woad (Isatis tinctoria) for possible commercial production of woad indigo. J. Agric. Eng. Res. 7: 315–320.
Tatusov R.L., Galperin M.Y., Natale D.A. & Koonin E.V. 2000. The COG database: a tool for genomescale analysis of protein functions and evolution. Nucleic Acids Res. 28: 33–36.
Thudi M., Li Y., Jackson S.A., May G.D. & Varshney R.K. 2012. Current stateofart of sequencing technologies for plant genomic research. Brief. Funct. Genomics 11: 3–11.
TroncosoPonce M.A., Kilaru A., Cao X., Durrett T.P., Fan J., Jensen J.K., Thrower N.A., Pauly M., Wilkerson C. & Ohlrogge J.B. 2011. Comparative deep transcriptional pro filing of four developing oilseeds. Plant J. 68: 1014–1027.
Ye J., Fang L., Zheng H.K., Zhang Y., Chen J., Zhang Z., Wang J., Li S., Li R., Bolund L. & Wang J. 2006. WEGO: a web tool for plotting GO annotations. Nucleic Acids Res. 34: W293–W297.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Minami, Y., Sarangi, B.K. & Thul, S.T. Transcriptome analysis for identification of indigo biosynthesis pathway genes in Polygonum tinctorium. Biologia 70, 1026–1032 (2015). https://doi.org/10.1515/biolog-2015-0131
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1515/biolog-2015-0131