Abstract
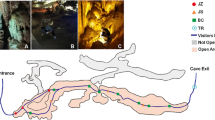
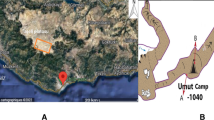
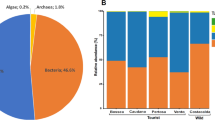
In our work, microbial diversity of Krubera-Voronja cave was evaluated in the view of the frequency of human visits in different locations as well as the sampling depth. Sampling in this cave was performed at depths of 220 m to 1640 m. Cultivation-independent method, namely barcoded pyrosequencing of 16S rRNA gene, was used for this analysis. Our results demonstrated high bacterial diversity at the phylum and genus levels. We have shown that the bacterial diversity at the phylum level depends on both the sampling depth and the frequency of human visits in Krubera-Voronja cave. Frequently visited locations were more diverse at the phylum level than the rarely visited branches. The total number of bacterial genera both per phylum and per sample correlated with the frequency of human visits but not with the sampling depth. Some genera, found in Krubera-Voronja cave, seem to be absent or very rare in other caves. The present study represents the first report on the microbial diversity in Krubera-Voronja cave.
Similar content being viewed by others
References
Barton H.A., Taylor M.R. & Pace N.R. 2004. Molecular phylogenetic analysis of a bacterial community in an oligotrophic cave environment. Geomicrobiol. J. 21: 11–20.
Bastian R., Alabouvette C. & Saiz-Jimenez C. 2009. Bacteria and free-living amoeba in the Lascaux cave. Res. Microbiol. 160: 38–40.
Carmichael M.J., Carmichael S.K., Santelli C.M., Strom A. & Bräuer S.L. 2013. Mn(II)-oxidizing bacteria are abundant and environmentally relevant members of ferromanganese deposits in caves of the Upper Tennessee River Basin. Geomicrobiol. J. 30: 779–800.
Chakravorty S., Helb D., Burday M., Connell N. & Alland D. 2007. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 69: 330–339.
Chan K.G. & Chong T.M. 2014. Prevalence of unclassified bacteria in tropical coastal waters of Malaysia revealed by metagenomic approach. Genome Announc. 2: e00419-14.
Clarke K.R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18: 117–143.
Cole J.R., Wang Q., Fish J.A., Chai B., McGarrell D.M., Sun Y., Brown C.T., Porras-Alfaro A., Kuske C.R. & Tiedje J.M. 2014. Ribosomal Database Project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. 42: D633–D642.
Cuezva S., Fernandez-Cortes A., Porca E., Pašić L., Jurado V., Hernandez-Marine M., Serrano-Ortiz P., Hermosin B., Cañaveras J.C., Sanchez-Moral S. & Saiz-Jimenez C. 2012. The biogeochemical role of Actinobacteria in Altamira Cave, Spain. FEMS Microbiol. Ecol. 81: 281–290.
Dillies M.A., Rau A., Aubert J., Hennequet-Antier C., Jeanmougin M., Servant N., Keime C., Marot G., Castel D., Estelle J., Guernec G., Jagla B., Jouneau L., Laloë D., Le Gall C., Schaëffer B., Le Crom S., Guedj M. & Jaffrézic F. 2013. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief. Bioinformatics 14: 671–683.
Edgar R.C., Haas B.J., Clemente J.C., Quince C. & Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27: 2194–2200.
Engei A.S. 2010. Microbial diversity of cave ecosystems, pp. 219–238. In: Loy A., Mandl M. & Barton L. (eds), Geomicro-biology: Molecular and Environmental Perspective, Springer Science+Business Media B.V., Dordrecht.
Engel A.S., Paoletti M.G., Beggio M., Dorigo L., Pamio A., Gomiero T., Furlan C., Brilli M., Dreon A.L., Bertoni R. & Squartini A. 2013. Comparative microbial community composition from secondary carbonate (moonmilk) deposits: implications for the Cansiliella servadeii cave hygropetric food web. Int. J. Speleol. 42: 181–192.
Epure L., Meleg I.N., Munteanu C.-M., Roban R.D. & Moldovan O.T. 2014. Bacterial and fungal diversity of quaternary cave sediment deposits. Geomicrobiol. J. 31: 116–127.
Gan H.Y., Gan H.M., Tarasco A.M., Busairi N.I., Barton H.A., Hudson A.O. & Savka M.A. 2014. Whole-genome sequences of five oligotrophic bacteria isolated from deep within Lechuguilla Cave, New Mexico. Genome Announc. 2: e01133-14.
Griffin D.W., Gray M.A., Lyles M.B. & Northup D.E. 2014. The transport of nonindigenous microorganisms into caves by human visitation: a case study at Carlsbad Caverns National Park. Geomicrobiol. J. 31: 175–185.
Groth I., Schumann P., Laiz L., Sanchez-Moral S., Cahveras J.C. & Saiz-Jimenez C. 2001. Geomicrobiological study of the Grotta dei Cervi, Porto Badisco, Italy. Geomicrobiol. J. 18: 241–258.
Jones D.S., Schaperdoth I. & Macalady J.L. 2014. Metage-nomic evidence for sulfide oxidation in extremely acidic cave biofilms. Geomicrobiol. J. 31: 194–204.
Jurado V., Laiz L., Rodriguez-Nava V., Boiron P., Boiron P., Hermosin B., Sanchez-Moral S. & Saiz-Jimenez C. 2010. Pathogenic and opportunistic microorganisms in caves. Int. J. Speleol. 39: 15–24.
Klindworth A., Pruesse E., Schweer T., Peplies J., Quast C., Horn M. & Glockner F.O. 2013. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 41: e1.
Kodama Y., Shumway M. & Leinonen R. 2012. The sequence read archive: explosive growth of sequencing data. Nucleic Acids Res. 40 (Database Issue): D54–D56.
Kumaresan D., Wischer D., Stephenson J., Hillebrand-Voiculescu A. & Murrell J.C. 2014. Microbiology of Movile Cave a chemolithoautotrophic ecosystem. Geomicrobiol. J. 31: 186–193.
McLellan S.L. & Eren A.M. 2014. Discovering new indicators of fecal pollution. Trends Microbiol. 22: 697–706.
Northup D.E. & Lavoie K.H. 2001. Geomicrobiology of caves: a review. Geomicrobiol. J. 18: 199–222.
Northup D.E., Melim L.A., Spilde M.N., Hathaway J.J.M., Garcia M.G., Moya M., Stone F.D., Boston P.J., Dapkevicius M.L.N.E. & Riquelme C. 2011. Lava cave microbial communities within mats and secondary mineral deposits: implications for life detection on other planets. Astrobiology 11: 601–618.
Ortiz M., Neilson J.W., Nelson W.M., Legatzki A., Byrne A., Yu Y., Wing R.A., Soderlund C.A., Pryor B.M., Pierson III L.S. & Maier R.M. 2013. Profiling bacterial diversity and taxonomic composition on speleothem surfaces in Kartchner Caverns, AZ. Microb. Ecol. 65: 371–383.
Porat L., Vishnivetskaya T.A., Mosher J.J., Brandt C.C., Yang Z.K., Brooks S.C., Liang L., Drake M.M., Podar M., Brown S.D., Palumbo A.V. 2010. Characterization of archaeal community in contaminated and uncontaminated surface stream sediments. Microb. Ecol. 60: 784–795.
Porter M.L., Engel A.S., Kane T.C. & Kinkle B.K. 2009. Productivity-diversity relationships from chemolithoautotrophically based sulfidic karst systems. Int. J. Speleol. 38: 27–40.
Portillo M.C., Gonzalez J.M. & Saiz-Jimenez C. 2008. Metabolically active microbial communities of yellow and grey colonizations on the walls of Altamira Cave, Spain. J. Appl. Microbiol. 104: 681–691.
Portillo M.C., Saiz-Jimenez C. & Gonzalez J.M. 2009. Molecular characterization of total and metabolically active bacterial communities of “white colonizations’ in the Altamira Cave, Spain. Res. Microbiol. 160: 41–47.
Schabereiter-Gurtner C., Saiz-Jimenez C., Pińar G., Lubitz W. & Rolleke S. 2002. Altamira cave Paleolithic paintings harbor partly unknown bacterial communities. FEMS Microbiol. Lett. 211: 7–11.
Sendra A. & Reboleira A.S.P.S. 2012. The world’s deepest subterranean community Krubera-Voronja Cave (Western Caucasus). Int. J. Speleol. 41: 221–230.
Studholme D.J., Jackson R.A. & Leak D.J. 1999. Phylogenetic analysis of transformable strains of thermophilic Bacillus species. FEMS Microbiol. Lett. 172: 85–90.
Acknowledgements
This work was supported by the Research Council of Lithuania (grant No. MIP-005/2014) and the National Geographic’s Global Exploration Fund (grant No. GEFNE50-Y12). Lithuanian caving club ‘Aenigma’ and Ukrainian Speleological Association are acknowledged for their cooperation in sampling. We would like to express our gratitude to Vytautas Zurauskas for his valuable consultations and assistance with bioinformatics.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kieraite-Aleksandrova, I., Aleksandrovas, V. & Kuisiene, N. Down into the Earth: microbial diversity of the deepest cave of the world. Biologia 70, 989–1002 (2015). https://doi.org/10.1515/biolog-2015-0127
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1515/biolog-2015-0127