Abstract
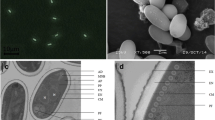
A new microsporidium, Nosema sp. PM-1, was first isolated from Papilio machaon Linnaeus. The spore shape of the PM-1 isolate was a long oval with an average size of 3.22 μm × 1.96 μm. Ultrastructure observation showed that PM-1 had a typical Nosema common diplokaryotic nuclei structure with 10–13 polar filament coils, spore wall, plasma membrane, and anchoring disk. The complete rRNA gene sequences were obtained by polymerase chain reaction amplification and each rRNA unit was arrayed as follows: 5′-LSU (2497 bp)-ITS (179 bp)-SSU (1232 bp)-IGS (278 bp)-5S (115 bp)-3′, which was the same as typical Nosema. The phylogenetic trees of rRNA, DNA-directed RNA polymerase II subunit, and tubulin genes all show that PM-1 was a sister to the clade comprising Nosema bombycis, Nosema spodopterae, and Nosema sp. PX1. The spore morphology, ultrastructure, and complete rRNA structure indicate that this isolate assigned to the “true” Nosema group, can parasitized in Papilio machaon Linnaeus, which provides a wider host range for Nosema.
Similar content being viewed by others
References
Balbiani G. 1882. Sur les microsporides ou psorospermies des Articules. Comptes rendus de l’Académie des Sciences, Paris, 95, 1168–1171
Canning E.U., Curry A., Cheney S., Lafranchi-Tristem N.J., Haque M.A. 1999. Vairimorpha imperfect a N. sp., a microsporidian exhibiting an abortive octosporous sporogony in Plutellaxylostella L. (Lepidoptera: Yponomeutidae). Parasitology, 119, 273–286
Choi J.Y., Kim J.G., Choi Y.C., Goo T.W., Chang J.H., Je Y.H., Kim K.Y. 2002. Nosema sp. isolated from cabbage white butterfly (Pieris rapae) collected in Korea. The Journal of Microbiology, 40, 199–204
Dong S.N., Shen Z.Y., Zhu F., Xu D.T., Xu L. 2011. Complete sequence and gene organization of the Nosema heliothidis ribosomal RNA gene region. Journal of Eukaryotic Microbiology, 58, 539–541. DOI: 10.1111/j.1550-7408.2011.00579.x
Edlind T.D., Li J., Visvesvara G.S., Vodkin M.H., Mclaughlin G.L., Katiyar S. 1996. Phylogenetic analysis of beta-tubulin sequences from a mitochondrial protozoa. Molecular Phylogenetics and Evolution, 5, 359–367. DOI: 10.1006/mpev.1996.0031
Hsu T.H., Hsu E.L., Yen F.Y. 1991. Spore ultrastructure of a microsporidian species (Nosema sp.) from the tobacco cutworm, Spodoptera litura. Chinese Journal of Entomology, 11, 242–251 (in Chinese with English summary)
Hsu T.H., Hsu E.L., Yen D.F. 1992. Nosema spodopterae new species, a new species of microsporidia from the tobacco cutworm, Spodoptera litura. Journal of the Agricultural Association of China, 157, 81–90 (in Chinese with English summary)
Huang W.F., Tsai S.J., Lo C.F., Soichi Y., Wang C.H. 2004. The novel organization and complete sequence of the ribosomal gene of Nosema bombycis. Fungal Genetics and Biology, 41, 473–481. DOI: 10.1016/j.fgb.2003.12.005.
Keeling P.J. 2003. Congruent evidence from alpha-tubulin and betatubulin gene phylogenies for a zygomycete origin of microsporidia. Fungal Genetics and Biology, 38, 298–309. DOI: 10.1016/S1087-1845(02)00537-6
Keeling P.J. 2009. Five Questions about Microsporidia. Public Library of Science (PLoS) Pathogens, 5(9), e1000489. DOI: 10.1371/journal.ppat.1000489
Ku C.T. 2004. The characteristics of two Nosema isolates (Nosema sp. PX1 & 2) from Plutellaxylostella in Taiwan. MD Thesis, National Taiwan University, Taiwan, China
Ku C.T., Wang C.Y., Tsai Y.C., Tzeng C.C., Wang C.H. 2007. Phylogenetic analysis of two putative Nosema isolates from cruciferous lepidopteron pests in Taiwan. Journal of Invertebrate Pathology, 95, 71–76. DOI: 10.1016/j.jip. 2006.11.008
Lee R.C., Williams B.A., Brown A.M., Adamson M.L., Keeling P.J., Fast N.M. 2008. Alpha- and beta-tubulin phylogenies support a close relationship between the microsporidia Brachiola algerae and Antonospora locustae. Journal of Eukaryotic Microbiology, 55, 388–392. DOI: 10.1111/j.1550-7408.2008. 00348.x
Liu H.D., Pan G.Q., Song S.H., Xu J.S.H., Li T., Deng Y.B., Zhou Z.Y. 2008. Multiple rDNA units distributed on all chromosomes of Nosema bombycis. Journal of Invertebrate Pathology, 99, 235–238. DOI: 10.1016/j.jip.2008.06.012
Liu H.D., Pan G.Q., Li T., Huang W., Luo B., Zhou Z.Y. 2012. Ultrastructure, chromosomal karyotype, and molecular phylogeny of a new isolate of microsporidian Vairimorpha sp. BM (Microsporidia, Nosematidae) from Bombyx mori in China. Parasitology Research, 110, 205–210. DOI: 10.1007/s00436-011-2470-9
Liu T., Xu J.SH., Li T., Pan G.Q., He Q., Li X.Y., Zhou Z.Y. 2014. Identification and Phylogenetic Analysis of MITEs families in a Microsporidia Isolated from Papilio machaon Linnaeus. Science of Sericulture, 40, 52–58
Nageli K., 1857. Uber die neue krankheit der seidenraupe und verwandte organismen. Botanische Zeitung, 15, 760–761
Pan G.Q. 2008. Studies on Nosema bombycis Genomics, Analyses of Genome Biology and Genome Evolution. PhD Thesis, Southwest university, Chongqing, China
Sprague V., Vavra J. 1977. Classification and phylogeny of the Microsporidia. In: (Eds. Bulla, L.A. and Cheng, T.C.) Comparative Pathobiology, Plenum Press, New York, 1–30
Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. 2011 MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Molecular Biology and Evolution, 28, 2731–2739. DOI: 10.1093/molbev/msr121
Tsai S.J., Huang W.F., Wang C.H. 2005. Complete sequence and gene organization of the Nosema spodopterae rRNA gene. Journal of Eukaryotic Microbiology, 52, 52–54. DOI: 10.1111/j.1550-7408.2005.3291rr.x
Tsai S.J., Lo C.F., Soichi Y., Wang C.H. 2003. The characterization of microsporidian isolates (Nosematidae: Nosema) from five important lepidopteran pests in Taiwan. Journal of Invertebrate Pathology, 83, 51–59. DOI: 10.1016/S0022-2011(03) 00035-1
Tsai Y.C., Solter F.L., Wang C.Y., Fan H.S., Chang C.C., Wang C.H. 2009. Morphological and molecular studies of a microsporidium (Nosema sp.) isolated from the three spot grass yellow butterfly, Eurema blanda arsakia(Lepidoptera: Pieridae). Journal of Invertebrate Pathology, 100, 85–93. DOI: 10.1016/j.jip.2008.11.006
Xu J.SH., Zhou Z.Y. 2010. Improving phylogenetic inference of microsporidian Nosema antheraeae. Journal of Biotechnology, 9, 7900–7904
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Liu, T., Xu, J., Luo, B. et al. Nosema sp. PM-1, a new isolate of microsporidian from infected Papilio machaon Linnaeus, based on ultrastructure and molecular identification. Acta Parasit. 60, 330–336 (2015). https://doi.org/10.1515/ap-2015-0046
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1515/ap-2015-0046