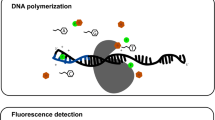
Abstract
Single-nucleotide polymorphisms (SNPs) are increasingly used as genetic markers. Although a high number of SNP-genotyping techniques have been described, most techniques still have low throughput or require major investments. For laboratories that have access to an automated sequencer, a single-base extension (SBE) assay can be implemented using the ABI SNaPshot™ kit. Here we present a modified protocol comprising multiplex template generation, multiplex SBE reaction, and multiplex sample analysis on a gel-based sequencer such as the ABI 377. These sequencers run on a Macintosh platform, but on this platform the software available for analysis of data from the ABI 377 has limitations. First, analysis of the size standard included with the kit is not facilitated. Therefore a new size standard was designed. Second, using Genotype (ABI), the analysis of the data is very tedious and time consuming. To enable automated batch analysis of 96 samples, with 10 SNPs each, we developed SNPtyper. This is a spreadsheet-based tool that uses the data from Genotyper and offers the user a convenient interface to set parameters required for correct allele calling. In conclusion, the method described will enable any lab having access to an ABI sequencer to genotype up to 1000 SNPs per day for a single experimenter, without investing in new equipment.
Similar content being viewed by others
References
Dietrich, W. F., Weber, J. L., Nickerson, D. A., and Kwok, P. Y. (1999) Identification and analysis of DNA polymorphisms, in: Genome Analysis, a Laboratory Manual, vol. 4: Mapping Genomes (Green, E. D., Birren, B., Klapholz, S., Myers, R. M., and Hieter, P., eds.), Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 135–186.
Saiki, R. K., Walsh, P. S., Levenson, C. H., and Erlich, H. A. (1989) Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl. Acad. Sci. USA 86, 6230–6234.
Hacia, J. G., Sun, B., Hunt, N., et al. (1998) Strategies for mutational analysis of the large multiexon ATM gene using high-density oligonucleotide arrays. Genome Res. 8, 1245–1258.
Prince, J. A., Feuk, L., Howell, W. M., et al. (2001) Robust and accurate single nucleotide polymorphism genotyping by dynamic allele-specific hybridization (DASH): design criteria and assay validation. Genome Res. 11, 152–162.
Livak, K. J., Flood, S. J., Marmaro, J., Giusti, W., and Deetz, K. (1995) Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 4, 357–362.
Livak, K. J. (1999) Allelic discrimination using fluorogenic probes and the 5′ nuclease assay. Genet. Anal. 14, 143–149.
Tyagi, S. and Kramer, F. R. (1996) Molecular beacons: probes that fluoresce upon hybridization. Nat. Biotechnol. 14, 303–308.
Pastinen, T., Kurg, A., Metspalu, A., Peltonen, L., and Syvanen, A. C. (1997) Minisequencing: a specific tool for DNA analysis and diagnostics on oligonucleotide arrays. Genome Res. 7, 606–614.
Nikiforov, T. T., Rendle, R. B., Goelet, P., et al. (1994) Genetic bit analysis: a solid phase method for typing single nucleotide polymorphisms. Nucleic Acids Res. 22, 4167–4175.
Braun, A., Little, D. P., and Koster, H. (1997) Detecting CFTR gene mutations by using primer oligo base extension and mass spectrometry. Clin. Chem. 43, 1151–1158.
Kristensen, V. N., Kelefiotis, D., Kristensen, T., and Børresen-Dal, A. L. (2001) High-throughput methods for detection of genetic variation. Biotechniques 30, 318–332.
Syvänen, A.-C. (2001) Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genetics 2, 930–942.
Syvänen, A.-C. (1999) From gels to chips: “minisequencing” primer extension for analysis of point mutations and single nucleotide polymorphisms. Hum. Mutat. 13, 1–10.
Lindblad-Toh, K., Winchester, E., Daley, M. J., et al. (2000) Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24, 381–386.
Henegariu, O., Heerema, N. A., Dlouhy, S. R., Vance, G. H., and Vogt, P. H. (1997) Multiplex PCR: critical parameters and step-by-step protocol. Biotechniques 23, 504–511.
Makridakis, N. M. and Reichardt, J. K. (2001) Multiplex automated primer extension analysis: simultaneous genotyping of several polymorphisms. Biotechniques 31 1374–1380.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jungerius, B.J. Typing single-nucleotide polymorphisms using a gel-based sequencer. Mol Biotechnol 25, 283–287 (2003). https://doi.org/10.1385/MB:25:3:283
Issue Date:
DOI: https://doi.org/10.1385/MB:25:3:283