Abstract
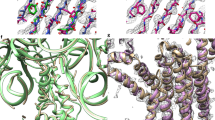
We propose a new approach for calculating the three-dimensional (3D) structure of a protein from distance and dihedral angle constraints derived from experimental data. We suggest that such constraints can be obtained from experiments such as tritium planigraphy, chemical or enzymatic cleavage of the polypeptide chain, paramagnetic perturbation of nuclear magnetic resonance (NMR) spectra, measurement of hydrogen-exchange rates, mutational studies, mass spectrometry, and electron paramagnetic resonance. These can be supplemented with constraints from theoretical prediction of secondary structures and of buried/exposed residues. We report here distance geometry calculations to generate the structures of a test protein Staphylococcal nuclease (STN), and the HIV-1 rev protein (REV) of unknown structure. From the available 3D atomic coordinates of STN, we set up simulated data sets consisting of varying number and quality of constraints, and used our group's Self Correcting Distance Geometry (SECODG) program DIAMOD to generate structures. We could generate the correct tertiary fold from qualitative (approximate) as well as precise distance constraints. The root mean square deviations of backbone atoms from the native structure were in the range of 2.0 Å to 8.3 Å, depending on the number of constraints used. We could also generate the correct fold starting from a subset of atoms that are on the surface and those that are buried. When we used data sets containing a small fraction of incorrect distance constraints, the SECODG technique was able to detect and correct them. In the case of REV, we used a combination of constraints obtained from mutagenic data and structure predictions. DIAMOD generated helix-loop-helix models, which, after four self-correcting cycles, populated one family exclusively. The features of the energy-minimized model are consistent with the available data on REV-RNA interaction. Our method could thus be an attractive alternative for calculating protein 3D structures, especially in cases where the traditional methods of X-ray crystallography and multidimensional NMR spectroscopy have been unsuccessful.
Similar content being viewed by others
References
Bogacheva, E. N., Gol'danskii, V. I., Shishkov A. V., Galkin, A. V., and Baratova, L. A. (1998) Tritium planigraphy: from the accessible surface to the spatial structure of a protein. Proc. Natl. Acad. Sci. USA 95, 2790–2794.
Volynskaya, A. V., Kasumov, E. A., Bogascheva, E. N., Shishkov, A. V., and Goldanskii, V. I. (1994) Determination of the accessible surface of globular proteins by means of tritium planigraphy. Eur. Biophys. J. 23, 139–143.
Esposito, G., Lesk, A. M., Molinari, H., Motta, A., Niccolai, N., and Pastore, A. (1992) Probing protein structure by solvent perturbation of nuclear magnetic resonance spectra. Nuclear magnetic resonance spectral editing and topological mapping in proteins by paramagnetic relaxation filtering. J. Mol. Biol. 224, 659–670.
Esposito, G., Lesk, A. M., Molinari, H., Motta, A., Niccolai, N., and Pastore, A. (1993) Probing protein structure by solvent perturbation of NMR spectra. II. Determination of surface and buried residues in homologous proteins. Biopolymers 33, 839–846.
Improta, S., Molinari, H., Pastore, A., Consonni, R., and Zetta, L. (1995) Probing protein structure by solvent perturbation of NMR spectra: a comparison with photochemically induced dynamic nuclear polarization techniques applied to native α-lactalbumin. Eur. J. Biochem. 227, 78–86.
Molinari, H., Esposito, G., Ragona, L., Pegna, M., Niccolai, N., Brunne, R. M., et al. (1997) Probing protein structure by solvent perturbation of NMR spectra: the surface accessibility of bovine pancreatic trypsin inhibitor. Biophys. J. 73, 382–396.
Spera, S., Ikura, M., and Bax, A. (1991) Measurement of the exchange rates of rapidly exchanging amide protons: application to the study of calmodulin and its complex with a myosin light chain kinase fragment. J. Biomol. NMR. 1, 155–165.
Mandell, J. G., Falick, A. M., and Komives, E. A. (1998) Measurement of amide hydrogen exchange by MALDI-TOF mass spectrometry. Anal. Chem. 70, 3987–3995.
Ermacora, M. R., Ledman, D. W., Hellinga, H. W., Hsu, G. W., and Fox, R. O. (1994) Mapping staphylococcal nuclease conformation using an EDTA-Fe derivative attached to genetically engineered cysteine residues. Biochemistry 33, 13,625–13,641.
Mumenthaler, C. and Braun, W. (1995) Predicting the helix packing of globular proteins by self-correcting distance geometry. Protein Sci. 4, 863–871.
Clore, G. M. and Gronenborn, A. M. (1999) Determining structures of large proteins and protein complexes by NMR, in Modern Techniques in Protein NMR, vol. 16 (Krishna, N. R., Berliner, L. J., eds.), Kluwer Academic/Plenum Publishers, New York, pp.
Wang, Y. X., Jacob, J., Cordier, F., Wingfield, P., Stahl, S. J., Lee-Huan, S., et al. (1999) Measurement of 3hJNC' connectivities across hydrogen bonds in a 30 kDa protein.
Cordier, F. and Grzesiek, S. (1999) Direct observation of hydrogen bonds in proteins by interresidue 3hJNC' scalar couplings. J. Am. Chem. Soc. 121, 1601–1602.
Zhu, H., Schein, C. H., and Braun, W. (2000) MASIA: recognition of patterns and properties in multiple aligned sequences. Bioinformatics In press.
Soman, K. V., Schein, C. H., Zhu, H., and Braun, W. (2000) Homology modeling and simulations of nuclease structures, in Nuclease Methods and Protocols, vol. 60 (Schein, C. H., ed.), Humana Press, Totowa, NJ.
Hanggi, G. and Braun, W. (1994) Pattern recognition and self-correcting distance geometry calculations applied to myohemerythrin. FEBS Lett. 344, 147–153.
Simons, K. T., Bonneau, R., Riczinski, I., and Baker, D. (1999) Ab initio protein structure prediction of CASP III targets using Rosetta. Prot. Struct. Funct. Gen. Suppl. 3, 171–176.
Sternberg, M. J. E., Bates, P. A., Kelley, L. A., and MacCallum, R. M. (1999) Progress in protein structure prediction: assessment of CASP3. Curr. Opin. Struct. Biol. 9, 368–373.
Günter, P., Braun, W., and Wüthrich, K. (1991) Efficient computation of three-dimensional protein structures in solution from NMR data using the program DIANA and the supporting programs CALIBA, HABAS, and GLOMSA. J. Mol. Biol. 217, 517–530.
Zhu, H. and Braun, W. (1999) Sequence specificity, statistical potentials and three-dimensional structure prediction with self-correcting distance geometry calculations of beta-sheet formation. Proc. Sci. 8, 1–17.
Hynes, T. R. and Fox, R. O. (1991) The crystal structure of staphylococcal nuclease refined at 1.7 Angstrom resolution. Prot. Struct. Funct. Genet. 10, 92.
Hope, T. J. (1999) The ins and outs of HIV. Rev. Arch. Biochem. Biophys. 365, 186–191.
Battiste, J. L., Mao, H., Rao, N. S., Tan, R., Muhandiram, D. R., Kay, L. E., et al. (1996) Alpha helix-RNA major groove recognition in an HIV-1 rev peptide-RRE RNA complex. Science 273, 1547–1551.
Scanlon, M. J., Fairlie, D. P., Craik, D. J., Englebretsen, D. R., and West, M. L. (1995) NMR solution structure of the RNA-binding peptide from human immunodeficiency virus (type 1) Rev. Biochem. 34, 8242–8249.
Ye, X., Gorin, A., Ellington, A. D., and Patel, D. J. (1996) Deep penetration of an alpha-helix into a widened RNA major groove in the HIV-1 rev peptide-RNA aptamer complex. Nat. Struct. Biol. 3, 1026–1033.
Auer, M., Gremlich, H.-U., Seifert, J.-M., Daly, T. J., Parslow, T. G., Casari, G., and Gstach, H. (1994) Helix-loop-helix motif in HIV-1 rev. Biochemistry 33, 2988–2996.
Watts, N. R., Misra, M., Wingfield, P. T., Stahl, S. J., Cheng, N., Trus, B. L., and Steven, A. C. (1998) Three-dimensional structure of HIV-1 rev protein filaments. J. Struct. Biol. 121, 41–52.
Thomas, S. L., Hauber, J., and Casari, G. (1997) Probing the structure of the HIV-1 Rev transactivator protein by functional analysis. Prot. Eng. 10, 103–107.
Thomas, S. L., Oft, M., Jaksche, H., Casari, G., Heger, P., Dobrovnik, M., et al. (1998) Functional analysis of the human immunodeficiency virus type 1 Rev protein oligomerization interface. J. Virol. 72, 2935–2944.
Fraczkiewicz, R. and Braun, W. (1998) Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comp. Chem. 19, 319–333.
Schaumann, T., Braun, W., and Wuthrich, K. (1990) The program FANTOM for energy refinement of polypeptides and proteins using a Newton-Raphson minimizer in torsion angle space. Biopolymers 29, 679–694.
Vila, J., Williams, R. L., Vasquez, M., and Scheraga, H. A. (1991) Empirical solvation models can be used to differentiate native from nearnative conformations of bovine pancreatic trypsin inhibitor. Prot. Struct. Funct. Gen. 10, 199–218.
Park, B. H., Huang, E. S., and Levitt, M. (1997) Factors affecting the ability of energy functions to discrimiate correct from incorrect folds. J. Mol. Biol. 266, 831–846.
Samudrala, R. and Moult, J. (1998) An all-atom distance dependent conditional probability discriminatory function for protein structure prediction. J. Mol. Biol. 275, 895–916.
Simons, K. T., Riczinski, I., Kooperberg, C., Fox, B. A., Bystroff, C., and Baker, D. (1999) Improved recognition of native-like protein structures using a combination of sequence-dependent and sequence-independent features of proteins. Proteins 34, 82–95.
Braun, W. (1987) Distance geometry and related methods for protein structure determination from NMR data. Q. Rev. Biophys. 19, 115–157.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Soman, K.V., Braun, W. Determining the three-dimensional fold of a protein from approximate constraints. Cell Biochem Biophys 34, 283–304 (2001). https://doi.org/10.1385/CBB:34:3:283
Issue Date:
DOI: https://doi.org/10.1385/CBB:34:3:283