Abstract
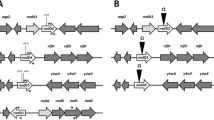
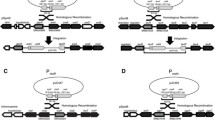
In Sinorhizobium meliloti, the nodD3 gene is transcriptionally controlled by two promoters, P1 and P2. Under P1, there is a 660 bp sequence including a small open reading frame, ORF2, followed by the nodD3 coding region. Genetic analysis using the different deletions on the 3’ ends of P1 downstream sequence showed that the downstream sequence +1–+125nt is essential for P1 expression. Complementation, mutations and nodulation tests demonstrated that the ORF2 auto-represses P1 expression, while the P1 downstream sequence +1–+125nt counteracts it.
Similar content being viewed by others
References
Fisher, R. F., Long, S. R., Rhizobium-plant signal exchange, Nature (London), 1992, 357: 55–660.
Schlaman, H. R. M., Okker, R. J. H., Lugtenberg, B. J. J., Regulation of nodulation gene expression by NodD in rhizobia, J. Bacteriol., 1992, 174: 5177–5182.
Denarie, J., Debelle, F., Rosenberg, C., Signaling and host range variation in nodulation, Annu. Rev. Microbiol., 1992, 46: 497–531.
Honma, M. A., Ausubel, F. M., Rhizobium meliloti has three functional copies of the nodD symbiotic regulatory gene, Proc. Natl. Acad. Sci. USA, 1987, 84: 8558–8562.
Kondorosi, E., Buire, M., Cren, M. et al., Involvement of the syrM and nodD3 genes of Rhizobium meliloti in nod gene activation and in optimal nodulation of the plant host, Mol. Microbiol., 1991, 5: 3035–3048.
Yu, G. Q., Zhu, J. B., Gu, J. et al., Evidence that the nodulation regulatory gene nodD3 of Rhizobium meliloti is transcribed from two separate promoters, Science in China, Ser. B, 1993, 36(3): 225–236.
Xiao, H., Shen, S. C., Zhu, J. B., Binding of activator SyrM to the site of nodD3 P1 region of Rhizobium meliloti, Science in China, Ser. C, 1998, 41(1): 2–6.
Kondorosi, E., Buire, M., Cren, M. et al., Involvement of the syrMand nodD3 genes of Rhizobium meliloti in nod gene activation and in optimal nodulation of the plant host, Mol. Mirobiol., 1991, 5: 3035–3048.
Dusha, I., Kondorosi, A., Genes at different regulatory levels are required for the ammonia control of nodulation in Rhizobium meliloti, Mol. Gen. Genet., 1993, 240: 435–444.
Dusha, I., Austin, S., Dixon, R., The upstream region of the nodD3 gene of Sinorhizobium meliloti carries enhancer sequences for the transcriptional activator NtrC, FEMS Microbiol. Lett., 1999, 179(2): 491–499.
Zhu, B., Dai, X. M., Zhu, J. B. et al., Regulatory role of the sequences downstream from nodD3 P1 promoter of Rhizobium meliloti, Chinese Science Bulletin, 2000, 45(1): 60–64.
Backman, K., Chen, Y. M., Magasanik, B., Physical and genetic characterization of the glnA-glnG region of the Escherichia coli chromosome, Proc. Natl. Acad. Sci. USA, 1981, 78: 3743–3747.
Ruvkun, G. B., Ausubel, F. M., A general method for site-directed mutagenesis in prokaryotes, Nature, 1981, 289: 85–89.
Meade, H. M., Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis, J. Bacteriol., 1982, 149: 114–122.
Yanisch-Perron, C., Vieira, J., Messing, J., Improved M13 cloning vectors and host strains: nucleotide sequence of the M13mp18 andpUC19 vectors, Gene, 1985, 33: 103–110.
Bagdasarian, M., Lurz, R., Ruckert, B. et al., Specific-purpose plasmid cloning vectors (II)—Broad host range, high copy number, RSF1010-derived vectors, and a host-vector system for gene cloning in Pseudomonas, Gene, 1981, 16: 237–247.
Spaink, H. P., Okler, R. J. H., Wijffelman, C. A. et al., Promoters in the nodulation region of the Rhizobium leguminosarum Symplasmid pRL1j1, Plant Mol. Biol., 1987, 9: 27–39.
Ditta, G., Stanfield, S., Corbin, D. et al., Broad host range DNA cloning system for Gram-negative bacteria: Construction of a gene bank of Rhizobium meliloti, Proc. Natl. Acad. Sci. USA, 1980, 77(12): 7347–7351.
Sambrook, J., Fritsh, E. F., Maniatis, T., Molecular Cloning: A Laboratory Manual, 2nd ed., New York: Cold Spring Harbor Laboratory Press, 1989.
Miller, J. H., Experiments in Molecular Genetics, New York: Cold Spring Harbor Laboratory Press, 1972.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chen, D., Liu, Y., Zhu, J. et al. Analysis of the downstream region of nodD3 P1 promoter by deletion and complementation tests in Sinorhizobium meliloti . Sci. China Ser. C.-Life Sci. 46, 165–173 (2003). https://doi.org/10.1360/03yc9018
Received:
Issue Date:
DOI: https://doi.org/10.1360/03yc9018