Abstract
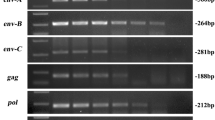
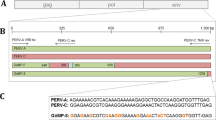
The provirus DNA sequence of porcine endogenous retrovirus (PERV) distributed in the pig genome is the major obstacle that restricts the swine as the organ donors in xenotransplantation, and the copy number of PERV varies greatly among different breeds and individuals. In the experiment, 67 healthy, female Chinese Experimental Mini-Pigs (CEMPs) aged at 3–6 months were selected from the Animal Husbandry Station of China Agricultural University, the copy number of PERV and types of envelope protein gene (env) were then investigated by means of PCR analysis and Southern blotting. It is showed that the distribution of types of envelope protein gene in Landrace and CEMPs makes little difference, but the proportion of individuals carrying two types of envelope protein gene (env-A and env-B, which is denoted as env-AB) is much larger than those which carry only one type of envelope protein gene (env-A or env-B). Meanwhile, two endogenous retrovirus free pigs were found for the first time during our research, and the copy number of others is relatively low, which is about 10 to 20. All the results illuminate the genetic diversity of indigenous pig breeds in China and the potential of CEMPs to serve as organ donors in xenotransplantation.
Similar content being viewed by others
References
Yin Zheng, Liu Jinghua, Animal Virology, Beijing: Science Press, 1997, 837–938.
Ward, T., Robert, M. P., Pamela, A. P., Role for β2-microglobulin in echovirus infection of rhobdomyosarcoma cells, J. Virol., 1998, 72(7): 5360–5365.
Heneine, W., Tibell, A., Switzer, W. M., Sandstrom, P., Rosales, G. V., Mathews, A., Korsgren, O., Chapman, L. E., Folks, T. M., Groth, C. G., No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts, Lancet, 1998, 352: 695–699.
Le Tissier, P., Stoye, J. P., Takeuchi, Y., Patience, C., Weiss, R. A., Two sets of human-tropic pig retrovirus, Nature, 1997, 389: 681–682.
Tonjess, R. R., Czauderna, F., Fisher, N., Molecularly cloned porcine endogenous retroviruses replicate on human cells, Transplantation Proceeding, 2000, 32(5): 1158–1161.
Lian Zhengxing, Rogel-Galliard, Li Ning, Patrick Chardon, Wu Changxin, Copy number polymorphism of endogenous retrovirus sequence in Chinese local pig breeds by Semi-PCR, Acta Veterinaria et Zootechnica Sinica, 2002, 33 (6): 521–524.
Teng Yuefeng, The richness of Chinese mini-pigs resources, World Agriculture, 1998, 10:41–43.
Rogel Gaillard, C., Bourgeaux, N., Billault, A., Vaiman, M., Chardon, P., Construction of a swine BAC library: Application to the characterization and mapping of porcine type C endoviral elements, Cytogenet. Cell Genet., 1999, 85: 205–211.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Lu, Q., Han, H., Lian, Z. et al. The screening and identification of endogenous retrovirus free CEMPs. Sci. China Ser. C.-Life Sci. 47, 562–566 (2004). https://doi.org/10.1360/03yc0181
Received:
Revised:
Issue Date:
DOI: https://doi.org/10.1360/03yc0181