Abstract
Background
The recurrence rate after hepatic resection of colorectal liver metastases (CRLM) remains high. This study aimed to investigate postoperative circulating tumor DNA (ctDNA) based on ultra-deep next-generation sequencing (NGS) to predict patient recurrence and survival.
Methods
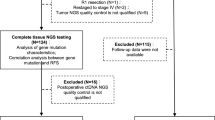
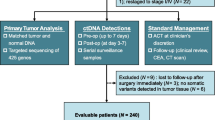
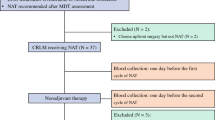
Using the high-throughput NGS method tagged with a dual-indexed unique molecular identifier, named the CRLM-specific 25-gene panel (J25), this study sequenced ctDNA in peripheral blood samples collected from 134 CRLM patients who underwent hepatectomy after postoperative day 6.
Results
Of 134 samples, 42 (31.3%) were shown to be ctDNA-positive, and 37 resulted in recurrence. Kaplan-Meier survival analysis showed that disease-free survival (DFS) in the ctDNA-positive subgroup was significantly shorter than in the ctDNA-negative subgroup (hazard ratio [HR], 2.96; 95% confidence interval [CI], 1.91–4.6; p < 0.05). When the 42 ctDNA-positive samples were further divided by the median of the mean allele frequency (AF, 0.1034%), the subgroup with higher AFs showed a significantly shorter DFS than the subgroup with lower AFs (HR, 1.98; 95% CI, 1.02–3.85; p < 0.05). The ctDNA-positive patients who received adjuvant chemotherapy longer than 2 months showed a significantly longer DFS than those who received treatment for 2 months or less (HR, 0.377; 95% CI, 0.189–0.751; p < 0.05). Uni- and multivariable Cox regression indicated two factors independently correlated with prognosis: ctDNA positivity and no preoperative chemotherapy.
Conclusion
The study demonstrated that ctDNA status 6 days postoperatively could sensitively and accurately predict recurrence for patients with CRLM using the J25 panel.





Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Adam R, Kitano Y. Multidisciplinary approach of liver metastases from colorectal cancer. Ann Gastroenterol Surg. 2019;3:50–6.
Margonis GA, Buettner S, Andreatos N, et al. Prognostic factors change over time after hepatectomy for colorectal liver metastases: a multi-institutional, international analysis of 1099 patients. Ann Surg. 2019;269:1129–37.
Imai K, Allard MA, Benitez CC, et al. Early recurrence after hepatectomy for colorectal liver metastases: what optimal definition and what predictive factors? Oncologist. 2016;21:887–94.
Naidoo M, Gibbs P, Tie J. ctDNA and adjuvant therapy for colorectal cancer: time to re-invent our treatment paradigm. Cancers. 2021;13:346.
To YH, Lee B, Wong HL, Gibbs P, Tie J. Circulating tumour DNA to guide treatment of gastrointestinal malignancies. Visc Med. 2020;36:388–96.
Alimirzaie S, Bagherzadeh M, Akbari MR. Liquid biopsy in breast cancer: a comprehensive review. Clin Genet. 2019;95:643–60.
Chabon JJ, Hamilton EG, Kurtz DM, et al. Integrating genomic features for non-invasive early lung cancer detection. Nature. 2020;580:245–51.
Kobayashi S, Nakamura Y, Taniguchi H, et al. Impact of preoperative circulating tumor DNA status on survival outcomes after hepatectomy for resectable colorectal liver metastases. Ann Surg Oncol. 2021;28:4744–55.
Mason MC, Tzeng CD, Tran Cao HS, et al. Preliminary analysis of liquid biopsy after hepatectomy for colorectal liver metastases. J Am Coll Surg. 2021;233:82–9.
Sing T, Sander O, Beerenwinkel N, Thomas L. ROCR: visualizing classifier performance in R. Bioinformatics. 2005; 21(20):3940–1. https://doi.org/10.1093/bioinformatics/bti623.
Tie J, Cohen JD, Wang Y, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019;68:663–71.
Fong Y, Fortner J, Sun RL, Brennan MF, Blumgart LH. Clinical score for predicting recurrence after hepatic resection for metastatic colorectal cancer: analysis of 1001 consecutive cases. Ann Surg. 1999;230:309–18 (discussion 318–21).
Tosi F, Magni E, Amatu A, et al. Effect of KRAS and BRAF mutations on survival of metastatic colorectal cancer after liver resection: a systematic review and meta-analysis. Clin Colorectal Cancer. 2017;16:e153–63.
Liu APY, Smith KS, Kumar R, et al. Serial assessment of measurable residual disease in medulloblastoma liquid biopsies. Cancer Cell. 2021;39:1519–30.
Diehl F, Schmidt K, Choti MA, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008;14:985–90.
Chen K, Zhao H, Shi Y, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res. 2019;25:7058–67.
Guo N, Lou F, Ma Y, et al. Circulating tumor DNA detection in lung cancer patients before and after surgery. Sci Rep. 2016;6:33519.
Loupakis F, Sharma S, Derouazi M, et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis Oncol. 2021;5:1166–77.
Warton K, Mahon KL, Samimi G. Methylated circulating tumor DNA in blood: power in cancer prognosis and response. Endocr Relat Cancer. 2016;23:R157–71.
Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 2019;5:1124–31.
Wang Y, Li L, Cohen JD, et al. Prognostic potential of circulating tumor DNA measurement in postoperative surveillance of nonmetastatic colorectal cancer. JAMA Oncol. 2019;5:1118–23.
Bidard FC, Kiavue N, Ychou M, et al. Circulating tumor cells and circulating tumor DNA detection in potentially resectable metastatic colorectal cancer: a prospective ancillary study to the Unicancer Prodige-14 Trial. Cells. 2019;8:516.
Narayan RR, Goldman DA, Gonen M, et al. Peripheral circulating tumor DNA detection predicts poor outcomes after liver resection for metastatic colorectal cancer. Ann Surg Oncol. 2019;26:1824–32.
Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016;8:346–92.
Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 2019;5:1710–7.
Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA. Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip. 2018;18:1174–96.
Mouliere F, Chandrananda D, Piskorz AM, et al. Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med. 2018;10:eaat4921.
Chakrabarti S, Xie H, Urrutia R, Mahipal A. The promise of circulating tumor DNA (ctDNA) in the management of early-stage colon cancer: a critical review. Cancers. 2020;12:2808.
Samorodnitsky E, Jewell BM, Hagopian R, et al. Evaluation of hybridization capture versus amplicon-based methods for whole-exome sequencing. Hum Mutat. 2015;36:903–14.
Garcia-Garcia G, Baux D, Faugere V, et al. Assessment of the latest NGS enrichment capture methods in clinical context. Sci Rep. 2016;6:20948.
Acknowledgment
We thank Hong-Wei Wang and Juan Li for their helpful suggestion in data analyzing. This study was supported by grants (nos. 81874143 and 31971192) from the National Nature Science Foundation of China and the Beijing Hospitals Authority Youth Program (code: QMS20201105).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Disclosure
There are no conflicts of interest.
Ethical Approval
The study was approved by the Ethics Committee of Beijing Cancer Hospital. The study was performed in accordance with the Declaration of Helsinki
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10434_2023_13362_MOESM4_ESM.pdf
The mutation oncoplot of tumor tissue sequenced by 642-gene panel in 134 patients. Genes in the J25 panel were highlighted in red. All tissue samples had at least one mutation detected in the genes of the J25 panel (PDF 26 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, W., Jin, KM., Zhang, MH. et al. Recurrence Prediction by Circulating Tumor DNA in the Patient with Colorectal Liver Metastases After Hepatectomy: A Prospective Biomarker Study. Ann Surg Oncol 30, 4916–4926 (2023). https://doi.org/10.1245/s10434-023-13362-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-023-13362-1