Abstract
Background
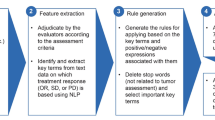
Accurate identification of pathologic complete response (pCR) from population-based electronic narrative data in a timely and cost-efficient manner is critical. This study aimed to derive and validate a set of natural language processing (NLP)-based machine-learning algorithms to capture pCR from surgical pathology reports of breast cancer patients who underwent neoadjuvant chemotherapy (NAC).
Methods
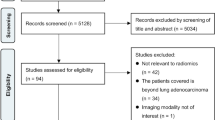
This retrospective cohort study included all invasive breast cancer patients who underwent NAC and subsequent curative-intent surgery during their admission at all four tertiary acute care hospitals in Calgary, Alberta, Canada, between 1 January 2010 and 31 December 2017. Surgical pathology reports were extracted and processed with NLP. Decision tree classifiers were constructed and validated against chart review results. Machine-learning algorithms were evaluated with a performance matrix including sensitivity, specificity, positive predictive value (PPV), negative predictive value [NPV], accuracy, area under the receiver operating characteristic curve [AUC], and F1 score.
Results
The study included 351 female patients. Of these patients, 102 (29%) achieved pCR after NAC. The high-sensitivity model achieved a sensitivity of 90.5% (95% confidence interval [CI], 69.6–98.9%), a PPV of 76% (95% CI, 59.6–87.2), an accuracy of 88.6% (95% CI, 78.7–94.9%), an AUC of 0.891 (95% CI, 0.795–0.987), and an F1 score of 82.61. The high-PPV algorithm reached a sensitivity of 85.7% (95% CI, 63.7–97%), a PPV of 81.8% (95% CI, 63.4–92.1%), an accuracy of 90% (95% CI, 80.5–95.9%), an AUC of 0.888 (95% CI, 0.790–0.985), and an F1 score of 83.72. The high-F1 score algorithm obtained a performance equivalent to that of the high-PPV algorithm.
Conclusion
The developed algorithms demonstrated excellent accuracy in identifying pCR from surgical pathology reports of breast cancer patients who received NAC treatment.



Similar content being viewed by others
References
Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
Waks AG, Winer EP. Breast cancer treatment: a review. JAMA. 2019;321:288–300.
Harbeck N, Penault-Llorca F, Cortes J, et al. Breast cancer. Nat Rev Dis Primers. 2019;5:1–31.
Dialani V, Chadashvili T, Slanetz PJ. Role of imaging in neoadjuvant therapy for breast cancer. Ann Surg Oncol. 2015;22:1416–24.
Mamounas EP. Impact of neoadjuvant chemotherapy on locoregional surgical treatment of breast cancer. Ann Surg Oncol. 2015;22:1425–33.
Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384:164–72.
Spring LM, Fell G, Arfe A, et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin Cancer Res. 2020;26:2838–48.
Pondé NF, Zardavas D, Piccart M. Progress in adjuvant systemic therapy for breast cancer. Nat Rev Clin Oncol. 2019;16:27–44.
Korn E, Sachs M, McShane L. Statistical controversies in clinical research: assessing pathologic complete response as a trial-level surrogate end point for early-stage breast cancer. Ann Oncol. 2016;27:10–5.
Cortazar P, Geyer CE. Pathological complete response in neoadjuvant treatment of breast cancer. Ann Surg Oncol. 2015;22:1441–6.
Locke S, Bashall A, Al-Adely S, et al. Natural language processing in medicine: a review. Trends Anaesth Crit Care. 2021;38:4–9.
Chowdhary K. Natural language processing. Fund Artif Intell. 2020. https://doi.org/10.1007/978-81-322-3972-7_19.
Duma N, Hoversten KP, Ruddy KJ. Exclusion of male patients in breast cancer clinical trials. JNCI Cancer Spect. 2018. https://doi.org/10.1093/jncics/pky018.
Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open. 2016;6:e012799.
Lee S, Xu Y, D’Souza AG, et al. Unlocking the potential of electronic health records for health research. Int J Population Data Sci. 2020. https://doi.org/10.23889/ijpds.v5i1.1123.
Pathological Complete Response in Neoadjuvant Treatment of High-Risk Early-Stage Breast Cancer: Use as an Endpoint to Support Accelerated Approval Guidance for Industry. Food and Drug Administration, 2020.
Honnibal M, Montani I. spaCy 2: natural language understanding with Bloom embeddings, convolutional neural networks and incremental parsing. To Appear. 2017;7:411–20.
Van Rossum, G., & Drake, F. L. (2009). Python 3 Reference Manual. Scotts Valley, CA: CreateSpace
Neumann M, King D, Beltagy I, et al. ScispaCy: fast and robust models for biomedical natural language processing. arXiv preprint arXiv: arXiv:190207669(2019).
A comparison of event models for I Bayes text classification. AAAI-98 workshop on learning for text categorization; 1998. Citeseer.
Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: machine learning in Python. J Mach Learn Res. 2011;12:2825–30.
Bekkar M, Djemaa HK, Alitouche TA. Evaluation measures for models assessment over imbalanced data sets. J Inf Eng Appl. 2013. https://doi.org/10.5121/ijdkp.2013.3402.
Breiman L, Friedman JH, Olshen RA, et al. Classification and regression trees. Routledge: New York, NY, 2017.
Mercaldo ND, Lau KF, Zhou XH. Confidence intervals for predictive values with an emphasis to case–control studies. Stat Med. 2007;26:2170–83.
van Rossum G. Python reference manual. Department of Computer Science [CS] 1995(R 9525).
Tang R, Ouyang L, Li C, et al. Machine learning to parse breast pathology reports in Chinese. Breast Cancer Res Treat. 2018;169:243–50.
Yala A, Barzilay R, Salama L, et al. Using machine learning to parse breast pathology reports. Breast Cancer Res Treat. 2017;161:203–11.
Cain EH, Saha A, Harowicz MR, et al. Multivariate machine learning models for prediction of pathologic response to neoadjuvant therapy in breast cancer using MRI features: a study using an independent validation set. Breast Cancer Res Treat. 2019;173:455–63.
Li F, Yang Y, Wei Y, et al. Deep learning-based predictive biomarker of pathological complete response to neoadjuvant chemotherapy from histological images in breast cancer. J Translat Med. 2021;19:1–13.
Qu YH, Zhu HT, Cao K, et al. Prediction of pathological complete response to neoadjuvant chemotherapy in breast cancer using a deep learning (DL) method. Thorac Cancer. 2020;11:651–8.
Sutton EJ, Onishi N, Fehr DA, et al. A machine learning model that classifies breast cancer pathologic complete response on MRI post-neoadjuvant chemotherapy. Breast Cancer Res. 2020;22:1–11.
Song Y-Y, Ying L. Decision tree methods: applications for classification and prediction. Shanghai Arch Psychiatry. 2015;27:130.
Myles AJ, Feudale RN, Liu Y, et al. An introduction to decision tree modeling. J Chemomet. 2004;18:275–85.
Ford E, Rooney P, Oliver S, et al. Identifying undetected dementia in UK primary care patients: a retrospective case-control study comparing machine-learning and standard epidemiological approaches. BMC Med Informat Decision Making. 2019;19:1–9.
Kim H-E, Kim HH, Han B-K, et al. Changes in cancer detection and false-positive recall in mammography using artificial intelligence: a retrospective, multireader study. Lancet Digital Health. 2020;2:e138–48.
Acknowledgment
This project, entitled “Building Pipeline to Transform Real-World Data to Evidence to Improve Cancer Care,” was supported by the Canadian Cancer Society (CCS).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
DISCLOSURE
The authors declare that they have are no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, G., Cheligeer, C., Brisson, AM. et al. A New Method of Identifying Pathologic Complete Response After Neoadjuvant Chemotherapy for Breast Cancer Patients Using a Population-Based Electronic Medical Record System. Ann Surg Oncol 30, 2095–2103 (2023). https://doi.org/10.1245/s10434-022-12955-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-022-12955-6