Abstract
Background
Colorectal cancers (CRCs) may be classified according to underlying genetic and epigenetic changes including microsatellite instability (MSI) and the CpG island methylator phenotype (CIMP). However the relevance of these molecular characteristics, which are being increasingly used to guide adjuvant therapy, has not been defined for metastatic disease. Since adjunct chemotherapy is designed to prevent or target metastases, molecular characteristics of metastatic disease are relevant. This study evaluates molecular differences between primary colorectal cancers and matched lymph node (LN) metastases.
Methods
An Institutional Review Board (IRB)-approved, prospectively maintained, frozen tissue biobank was queried for stage III CRCs previously analyzed for MSI and CIMP. Metastatic cancer-containing LNs from the same patients were retrieved from formalin-fixed paraffin-embedded (FFPE) tissues. DNA was isolated from matched primary tumors and LNs, tested for MSI and CIMP, and the results were compared.
Results
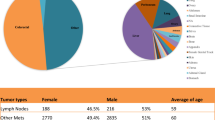
Forty-seven matched LNs from 47 CRC cases were available. Six of 47 primary tumors and 8/47 (17%) LNs were MSI-H (p = 0.25). Thirteen of 47 (28%) primary tumors and 6/47 (13%) LNs were CIMP+ (p < 0.02). Eight patients displayed nine disparities between their primary tumors and LNs: two for MSI and seven for CIMP. Interestingly, of the 13 CIMP+ primary tumors, seven had LN metastases that were CIMP negative.
Conclusions
Molecular characterization, notably the CpG island methylator phenotype, varies between primary tumors and corresponding lymphatic metastases. Although the mechanism for this is unknown, this finding suggests that molecular typing of LNs as well as primary tumors should be considered for molecular-based adjuvant therapy decisions.
Similar content being viewed by others
References
Jass JR. Classification of colorectal cancer based on correlation of clinical, morphological and molecular features. Histopathology. 2007;50:113–30.
Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–67.
Soreide K, Janssen EA, Soiland H, Korner H, Baak JP. Microsatellite instability in colorectal cancer. Br J Surg. 2006;93:395–406.
Suraweera N, Duval A, Reperant M, et al. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123:1804–11.
Bronner CE, Baker SM, Morrison PT, et al. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary non-polyposis colon cancer. Nature. 1994;368:258–61.
Leach FS, Nicolaides NC, Papadopoulos N, et al. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–25.
Samowitz WS, Albertsen H, Herrick J, et al. Evaluation of a large, population-based sample supports a CpG island methylator phenotype in colon cancer. Gastroenterology. 2005;129:837–45.
Aaltonen LA, Peltomaki P, Leach FS, et al. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–6.
Dumitrescu RG. Epigenetic targets in cancer epidemiology. Methods Mol Biol. 2009;471:457–67.
Kondo Y, Issa JP. Epigenetic changes in colorectal cancer. Cancer Metastasis Rev. 2004;23:29–39.
Toyota M, Ohe-Toyota M, Ahuja N, Issa JP. Distinct genetic profiles in colorectal tumors with or without the CpG island methylator phenotype. Proc Natl Acad Sci USA. 2000;97:710–5.
Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–96.
Baylin SB, Ohm JE. Epigenetic gene silencing in cancer—a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–16.
Kim YH, Petko Z, Dzieciatkowski S, et al. CpG island methylation of genes accumulates during the adenoma progression step of the multistep pathogenesis of colorectal cancer. Genes Chromosomes Cancer. 2006;45:781–9.
Boland CR, Goel A. Somatic evolution of cancer cells. Semin Cancer Biol. 2005;15:436–50.
Ogino S, Meyerhardt JA, Kawasaki T, et al. CpG island methylation, response to combination chemotherapy, and patient survival in advanced microsatellite stable colorectal carcinoma. Virchows Arch. 2007;450:529–37.
Ogino S, Nosho K, Kirkner GJ, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58:90–6.
Ribic CM, Sargent DJ, Moore MJ, et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N Engl J Med. 2003;349:247–57.
Shen L, Catalano PJ, Benson AB, III, O’Dwyer P, Hamilton SR, Issa JP. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res. 2007;13:6093–8.
Van Rijnsoever M, Elsaleh H, Joseph D, McCaul K, Iacopetta B. CpG island methylator phenotype is an independent predictor of survival benefit from 5-fluorouracil in stage III colorectal cancer. Clin Cancer Res. 2003;9:2898–903.
Kalady MF, Sanchez JA, Manilich E, Hammel J, Casey G, Church JM. Divergent oncogenic changes influence survival differences between colon and rectal adenocarcinomas. Dis Colon Rectum. 2009;52:1039–45.
de la Chapelle A. Testing tumors for microsatellite instability. Eur J Hum Genet. 1999;7:407–8.
Haddad R, Ogilvie RT, Croitoru M, et al. Microsatellite instability as a prognostic factor in resected colorectal cancer liver metastases. Ann Surg Oncol. 2004;11:977–82.
Eads CA, Danenberg KD, Kawakami K, et al. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32.
Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93.
Trinh BN, Long TI, Laird PW. DNA methylation analysis by MethyLight technology. Methods. 2001;25:456–62.
Chen WS, Chen JY, Liu JM, et al. Microsatellite instability in sporadic-colon-cancer patients with and without liver metastases. Int J Cancer. 1997;74:470–4.
Bird AP, Wolffe AP. Methylation-induced repression–belts, braces, and chromatin. Cell. 1999;99:451–4.
Drake JW. Comparative rates of spontaneous mutation. Nature. 1969;221:1132.
Aranda-Anzaldo A. Cancer development and progression: a non-adaptive process driven by genetic drift. Acta Biotheor. 2001;49:89–108.
Karpf AR, Jones DA. Reactivating the expression of methylation silenced genes in human cancer. Oncogene. 2002;21:5496–503.
Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:e157.
Scheel C, Onder T, Karnoub A, Weinberg RA. Adaptation versus selection: the origins of metastatic behavior. Cancer Res. 2007;67:11476–9; discussion 9–80.
Graff JR, Gabrielson E, Fujii H, Baylin SB, Herman JG. Methylation patterns of the E-cadherin 5’ CpG island are unstable and reflect the dynamic, heterogeneous loss of E-cadherin expression during metastatic progression. J Biol Chem. 2000;275:2727–32.
Wu JM, Fackler MJ, Halushka MK, et al. Heterogeneity of breast cancer metastases: comparison of therapeutic target expression and promoter methylation between primary tumors and their multifocal metastases. Clin Cancer Res. 2008;14:1938–46.
Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute workshop on microsatellite instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58:5248–57.
Pyatt R, Chadwick RB, Johnson CK, Adebamowo C, de la Chapelle A, Prior TW. Polymorphic variation at the BAT-25 and BAT-26 loci in individuals of African origin. Implications for microsatellite instability testing. Am J Pathol. 1999;155:349–53.
Samowitz WS, Slattery ML, Potter JD, Leppert MF. BAT-26 and BAT-40 instability in colorectal adenomas and carcinomas and germline polymorphisms. Am J Pathol. 1999;154:1637–41.
Sanchez J, Aung PP, Merkulova A, et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Cancer. 2009; accepted for publication.
Harbeck N, Nimmrich I, Hartmann A, et al. Multicenter study using paraffin-embedded tumor tissue testing PITX2 DNA methylation as a marker for outcome prediction in tamoxifen-treated, node-negative breast cancer patients. J Clin Oncol. 2008;26:5036–42.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Messick, C.A., Church, J.M., Liu, X. et al. Stage III Colorectal Cancer: Molecular Disparity Between Primary Cancers and Lymph Node Metastases. Ann Surg Oncol 17, 425–431 (2010). https://doi.org/10.1245/s10434-009-0783-z
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1245/s10434-009-0783-z