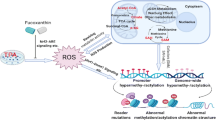
Abstract
Overexposure to ultraviolet radiation and environmental carcinogens drive skin cancer development through redox imbalance and gene mutation. Antioxidants such as triterpenoids have exhibited anti-oxidative and anti-inflammatory potentials to alleviate skin carcinogenesis. This study investigated the methylome and transcriptome altered by tumor promoter 12-O-tetradecanoylphorbol-13-acetate (TPA) or TPA with 2-cyano 2,3-dioxoolean-1,9-dien-28-oic acid (CDDO). The results show that CDDO blocks TPA-induced transformation dose dependently. Several differential expressed genes (DEGs) involved in skin cell transformation, while counteracted by CDDO, were revealed by differential expression analysis including Lyl1, Lad1, and Dennd2d. In CpG methylomic profiles, the differentially methylated regions (DMRs) in the promoter region altered by TPA while showing the opposite methylation status in the CDDO treatment group were identified. The correlation between DNA methylation and RNA expression has been established and DMRs showing inverse correlation were further studied as potential therapeutic targets. From the CpG methylome and transcriptome results, CDDO significantly restored gene expression of NAD(P)H:quinone oxidoreductase 1 (Nqo1) inhibited by TPA by decreasing their promoter CpG methylation. Ingenuity Pathways Analysis (IPA) shows that CDDO neutralized the effect of TPA through modulating cell cycles, cell migration, and inflammatory and immune response regulatory pathways. Notably, Tumor Necrosis Factor Receptor 2 (TNFR2) signaling was significantly downregulated by CDDO potentially contributing to prevention of TPA-induced cell transformation. Overall, incorporating the transcriptome, CpG methylome, and signaling pathway network, we reveal potential therapeutic targets and pathways by which CDDO could reverse TPA-induced carcinogenesis. The results could be useful for future human study and targets development for skin cancer.
Graphical Abstract







Similar content being viewed by others
References
Rogers HW, Weinstock MA, Feldman SR, Coldiron BM. Incidence estimate of nonmelanoma skin cancer (keratinocyte carcinomas) in the U.S. population, 2012. JAMA Dermatol. 2015;151(10):1081–6.
Aggarwal P, Knabel P, Fleischer AB. United States burden of melanoma and non-melanoma skin cancer from 1990 to 2019. J Am Acad Dermatol. 2021;85(2):388–95.
Madan V, Lear JT, Szeimies RM. Non-melanoma skin cancer. Lancet. 2010;375(9715):673–85. https://doi.org/10.1016/S0140-6736(09)61196-X.
Alam M, Ratner D. Cutaneous squamous-cell carcinoma. N Engl J Med. 2001;344(13):975–83. https://doi.org/10.1056/NEJM200103293441306.
Stewart BW, Kleihues P. World cancer report. Lyons: IARC Press; 2003. p. 232–6.
Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4(9):1350–62.
Khan AQ, Khan R, Qamar W, Lateef A, Rehman MU, Tahir M, et al. Geraniol attenuates 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced oxidative stress and inflammation in mouse skin: possible role of p38 MAP kinase and NF-kappaB. Exp Mol Pathol. 2013;94(3):419–29. https://doi.org/10.1016/j.yexmp.2013.01.006.
Kashyap MP, Sinha R, Mukhtar MS, Athar M. Epigenetic regulation in the pathogenesis of non-melanoma skin cancer. Semin Cancer Biol. 2022;83:36–56.
Saha K, Hornyak TJ, Eckert RL. Epigenetic cancer prevention mechanisms in skin cancer. AAPS J. 2013;15(4):1064–71.
Lubecka K, Kurzava L, Flower K, Buvala H, Zhang H, Teegarden D, et al. Stilbenoids remodel the DNA methylation patterns in breast cancer cells and inhibit oncogenic NOTCH signaling through epigenetic regulation of MAML2 transcriptional activity. Carcinogenesis. 2016;37(7):656–68.
Borella R, Forti L, Gibellini L, De Gaetano A, De Biasi S, Nasi M, et al. Synthesis and anticancer activity of CDDO and CDDO-Me, two derivatives of natural triterpenoids. Molecules. 2019;24(22):4097.
Du J-R, Long F-Y, Chen C. Chapter six – research progress on natural triterpenoid saponins in the chemoprevention and chemotherapy of cancer. In: Bathaie SZ, Tamanoi F, editors. The enzymes. Academic Press; 2014. p. 95–130.
Kim H, Ramirez CN, Su Z-Y, Kong A-NT. Epigenetic modifications of triterpenoid ursolic acid in activating Nrf2 and blocking cellular transformation of mouse epidermal cells. J Nutr Biochem. 2016;33:54–62.
Yang Y, Yin R, Wu R, Ramirez CN, Sargsyan D, Li S, et al. DNA methylome and transcriptome alterations and cancer prevention by triterpenoid ursolic acid in UVB-induced skin tumor in mice. Mol Carcinog. 2019;58(10):1738–53. https://doi.org/10.1002/mc.23046.
Hudlikar RR, Sargsyan D, Wu R, Su S, Zheng M, Kong AN. Triterpenoid corosolic acid modulates global CpG methylation and transcriptome of tumor promotor TPA induced mouse epidermal JB6 P+ cells. Chem Biol Interact. 2020;321:109025.
Liby KT, Yore MM, Sporn MB. Triterpenoids and rexinoids as multifunctional agents for the prevention and treatment of cancer. Nat Rev Cancer. 2007;7:357. https://doi.org/10.1038/nrc2129.
Li S, Kuo H-CD, Yin R, Wu R, Liu X, Wang L, et al. Epigenetics/epigenomics of triterpenoids in cancer prevention and in health. Biochem Pharmacol. 2020;175:113890.
Honda T, Rounds BV, Gribble GW, Suh N, Wang Y, Sporn MB. Design and synthesis of 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, a novel and highly active inhibitor of nitric oxide production in mouse macrophages. Bioorg Med Chem Lett. 1998;8(19):2711–4.
Suh N, Honda T, Finlay HJ, Barchowsky A, Williams C, Benoit NE, et al. Novel triterpenoids suppress inducible nitric oxide synthase (iNOS) and inducible cyclooxygenase (COX-2) in mouse macrophages. Cancer Res. 1998;58(4):717–23.
Suh N, Wang Y, Honda T, Gribble GW, Dmitrovsky E, Hickey WF, et al. A novel synthetic oleanane triterpenoid, 2-cyano-3,12-dioxoolean-1,9-dien-28-oic acid, with potent differentiating, antiproliferative, and anti-inflammatory activity. Cancer Res. 1999;59(2):336–41.
Suh N, Roberts AB, Birkey Reffey S, Miyazono K, Itoh S, ten Dijke P, et al. Synthetic triterpenoids enhance transforming growth factor beta/Smad signaling. Cancer Res. 2003;63(6):1371–6.
Place AE, Suh N, Williams CR, Risingsong R, Honda T, Honda Y, et al. The novel synthetic triterpenoid, CDDO-imidazolide, inhibits inflammatory response and tumor growth in vivo. Clin Cancer Res. 2003;9(7):2798–806.
To C, Ringelberg CS, Royce DB, Williams CR, Risingsong R, Sporn MB, et al. Dimethyl fumarate and the oleanane triterpenoids, CDDO-imidazolide and CDDO-methyl ester, both activate the Nrf2 pathway but have opposite effects in the A/J model of lung carcinogenesis. Carcinogenesis. 2015;36(7):769–81.
Liby K, Hock T, Yore MM, Suh N, Place AE, Risingsong R, et al. The synthetic triterpenoids, CDDO and CDDO-imidazolide, are potent inducers of heme oxygenase-1 and Nrf2/ARE signaling. Cancer Res. 2005;65(11):4789–98. https://doi.org/10.1158/0008-5472.CAN-04-4539.
Lapillonne H, Konopleva M, Tsao T, Gold D, McQueen T, Sutherland RL, et al. Activation of peroxisome proliferator-activated receptor gamma by a novel synthetic triterpenoid 2-cyano-3,12-dioxooleana-1,9-dien-28-oic acid induces growth arrest and apoptosis in breast cancer cells. Cancer Res. 2003;63(18):5926–39.
Yates MS, Kwak MK, Egner PA, Groopman JD, Bodreddigari S, Sutter TR, et al. Potent protection against aflatoxin-induced tumorigenesis through induction of Nrf2-regulated pathways by the triterpenoid 1-[2-cyano-3-,12-dioxooleana-1,9(11)-dien-28-oyl]imidazole. Cancer Res. 2006;66(4):2488–94. https://doi.org/10.1158/0008-5472.CAN-05-3823.
Deeb D, Brigolin C, Gao X, Liu Y, Pindolia KR, Gautam SC. Induction of apoptosis in pancreatic cancer cells by CDDO-Me involves repression of telomerase through epigenetic pathways. J Carcinog Mutagen. 2014;5:177.
Livingstone MC, Johnson NM, Roebuck BD, Kensler TW, Groopman JD. Profound changes in miRNA expression during cancer initiation by aflatoxin B1 and their abrogation by the chemopreventive triterpenoid CDDO-Im. Mol Carcinog. 2017;56(11):2382–90. https://doi.org/10.1002/mc.22635.
Kuo HD, Wu R, Li S, Yang AY, Kong AN. Anthocyanin delphinidin prevents neoplastic transformation of mouse skin JB6 P+ cells: epigenetic re-activation of Nrf2-ARE pathway. AAPS J. 2019;21(5):83.
Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–2.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60.
Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–30.
Wang L, Feng Z, Wang X, Wang X, Zhang X. DEGseq: an R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics. 2010;26(1):136–8.
Krueger F, Andrews SR. Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics. 2011;27(11):1571–2.
Gaspar JM, Hart RP. DMRfinder: efficiently identifying differentially methylated regions from MethylC-seq data. BMC Bioinformatics. 2017;18(1):528.
Yu G, Wang LG, He QY. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics. 2015;31(14):2382–3.
Zemach A, McDaniel IE, Silva P, Zilberman D. Genome-wide evolutionary analysis of eukaryotic DNA methylation. Science. 2010;328(5980):916. https://doi.org/10.1126/science.1186366.
Feng S, Cokus SJ, Zhang X, Chen P-Y, Bostick M, Goll MG, et al. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010;107(19):8689–94. https://doi.org/10.1073/pnas.1002720107.
Wei S-J, Trempus CS, Ali RC, Hansen LA, Tennant RW. 12-O-Tetradecanoylphorbol-13-acetate and UV radiation-induced nucleoside diphosphate protein kinase B mediates neoplastic transformation of epidermal cells. J Biol Chem. 2004;279(7):5993–6004.
Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4(9):1350.
Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol). 1995;57(1):289–300.
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci. 2005;102(43):15545–50. https://doi.org/10.1073/pnas.0506580102.
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27(12):1739–40. https://doi.org/10.1093/bioinformatics/btr260.
Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–25. https://doi.org/10.1016/j.cels.2015.12.004.
Mootha VK, Lindgren CM, Eriksson K-F, Subramanian A, Sihag S, Lehar J, et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–73. https://doi.org/10.1038/ng1180.
Konopleva M, Tsao T, Ruvolo P, Stiouf I, Estrov Z, Leysath CE, et al. Novel triterpenoid CDDO-Me is a potent inducer of apoptosis and differentiation in acute myelogenous leukemia. Blood. 2002;99(1):326–35.
Chintharlapalli S, Papineni S, Konopleva M, Andreef M, Samudio I, Safe S. 2-Cyano-3,12-dioxoolean-1,9-dien-28-oic acid and related compounds inhibit growth of colon cancer cells through peroxisome proliferator-activated receptor gamma-dependent and -independent pathways. Mol Pharmacol. 2005;68(1):119–28.
Yates MS, Tauchi M, Katsuoka F, Flanders KC, Liby KT, Honda T, et al. Pharmacodynamic characterization of chemopreventive triterpenoids as exceptionally potent inducers of Nrf2-regulated genes. Mol Cancer Ther. 2007;6(1):154–62.
Hyer ML, Shi R, Krajewska M, Meyer C, Lebedeva IV, Fisher PB, et al. Apoptotic activity and mechanism of 2-cyano-3,12-dioxoolean-1,9-dien-28-oic-acid and related synthetic triterpenoids in prostate cancer. Cancer Res. 2008;68(8):2927–33.
Ikeda T, Sporn M, Honda T, Gribble GW, Kufe D. The novel triterpenoid CDDO and its derivatives induce apoptosis by disruption of intracellular redox balance. Cancer Res. 2003;63(17):5551–8.
Guy GP Jr, Thomas CC, Thompson T, Watson M, Massetti GM, Richardson LC. Vital signs: melanoma incidence and mortality trends and projections – United States, 1982–2030. MMWR Morb Mortal Wkly Rep. 2015;64(21):591–6.
Guy GP Jr, Machlin SR, Ekwueme DU, Yabroff KR. Prevalence and costs of skin cancer treatment in the U.S., 2002–2006 and 2007–2011. Am J Prev Med. 2015;48(2):183–7. https://doi.org/10.1016/j.amepre.2014.08.036.
de Gruijl FR, van Kranen HJ, Mullenders LHF. UV-induced DNA damage, repair, mutations and oncogenic pathways in skin cancer. J Photochem Photobiol, B. 2001;63(1):19–27. https://doi.org/10.1016/S1011-1344(01)00199-3.
Fabbrocini G, Triassi M, Mauriello MC, Torre G, Annunziata MC, De Vita V, et al. Epidemiology of skin cancer: role of some environmental factors. Cancers (Basel). 2010;2(4):1980–9.
Chitsazzadeh V, Coarfa C, Drummond JA, Nguyen T, Joseph A, Chilukuri S, et al. Cross-species identification of genomic drivers of squamous cell carcinoma development across preneoplastic intermediates. Nat Commun. 2016;7:12601.
Hoang VLT, Tom LN, Quek XC, Tan JM, Payne EJ, Lin LL, et al. RNA-seq reveals more consistent reference genes for gene expression studies in human non-melanoma skin cancers. PeerJ. 2017;5:e3631.
San-Marina S, Han Y, Suarez Saiz F, Trus MR, Minden MD. Lyl1 interacts with CREB1 and alters expression of CREB1 target genes. Biochim Biophys Acta. 2008;1783(3):503–17. https://doi.org/10.1016/j.bbamcr.2007.11.015.
Nagel S, Venturini L, Meyer C, Kaufmann M, Scherr M, Drexler HG, et al. Multiple mechanisms induce ectopic expression of LYL1 in subsets of T-ALL cell lines. Leuk Res. 2010;34(4):521–8. https://doi.org/10.1016/j.leukres.2009.06.020.
Zhang M-M, Liu N, Zhang Y-L, Rong B, Wang X-L, Xu C-H, et al. Destabilization of AETFC through C/EBPα-mediated repression of LYL1 contributes to t(8;21) leukemic cell differentiation. Leukemia. 2019;33(7):1822–7. https://doi.org/10.1038/s41375-019-0398-8.
Fang F, Lu J, Sang X, Tao Y-F, Wang J-W, Zhang Z-M, et al. Super-enhancer profiling identifies novel critical and targetable cancer survival gene LYL1 in pediatric acute myeloid leukemia. J Exp Clin Cancer Res. 2022;41(1):225. https://doi.org/10.1186/s13046-022-02428-9.
Teixeira JC, de Filippo C, Weihmann A, Meneu JR, Racimo F, Dannemann M, et al. Long-term balancing selection in LAD1 maintains a missense trans-species polymorphism in humans, chimpanzees, and bonobos. Mol Biol Evol. 2015;32(5):1186–96. https://doi.org/10.1093/molbev/msv007.
Motoki K, Megahed M, LaForgia S, Uitto J. Cloning and chromosomal mapping of mouse ladinin, a novel basement membrane zone component. Genomics. 1997;39(3):323–30. https://doi.org/10.1006/geno.1996.4507.
Codreanu SG, Hoeksema MD, Slebos RJC, Zimmerman LJ, Rahman SMJ, Li M, et al. Identification of proteomic features to distinguish benign pulmonary nodules from lung adenocarcinoma. J Proteome Res. 2017;16(9):3266–76. https://doi.org/10.1021/acs.jproteome.7b00245.
Rusinek D, Swierniak M, Chmielik E, Kowal M, Kowalska M, Cyplinska R, et al. BRAFV600E-associated gene expression profile: early changes in the transcriptome, based on a transgenic mouse model of papillary thyroid carcinoma. PLoS ONE. 2015;10(12):e0143688. https://doi.org/10.1371/journal.pone.0143688.
Li J, Wang Z, Tie C. High expression of ladinin-1 (LAD1) predicts adverse outcomes: a new candidate docetaxel resistance gene for prostatic cancer (PCa). Bioengineered. 2021;12(1):5749–59. https://doi.org/10.1080/21655979.2021.1968647.
Roth L, Srivastava S, Lindzen M, Sas-Chen A, Sheffer M, Lauriola M, et al. SILAC identifies LAD1 as a filamin-binding regulator of actin dynamics in response to EGF and a marker of aggressive breast tumors. Sci Signal. 2018;11(515). https://doi.org/10.1126/scisignal.aan0949.
Uhlén M, Fagerberg L, Hallström Björn M, Lindskog C, Oksvold P, Mardinoglu A, et al. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419.
Human Protein Atlas proteinatlas.org. https://www.proteinatlas.org/ENSG00000159166-LAD1/pathology/skin+cancer. Gene available from v21.0. proteinatlas.org.
Uhlen M, Zhang C, Lee S, Sjöstedt E, Fagerberg L, Bidkhori G, et al. A pathology atlas of the human cancer transcriptome. Science. 2017;357(6352):eaan2507.
Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191(2):367–81. https://doi.org/10.1083/jcb.201008051.
Behnia R, Munro S. Organelle identity and the signposts for membrane traffic. Nature. 2005;438(7068):597–604. https://doi.org/10.1038/nature04397.
Zerial M, McBride H. Rab proteins as membrane organizers. Nat Rev Mol Cell Biol. 2001;2(2):107–17. https://doi.org/10.1038/35052055.
Hibino S, Kanda M, Oya H, Takami H, Shimizu D, Nomoto S, et al. Reduced expression of DENND2D through promoter hypermethylation is an adverse prognostic factor in squamous cell carcinoma of the esophagus. Oncol Rep. 2014;31(2):693–700. https://doi.org/10.3892/or.2013.2901.
Sakha S, Muramatsu T, Ueda K, Inazawa J. Exosomal microRNA miR-1246 induces cell motility and invasion through the regulation of DENND2D in oral squamous cell carcinoma. Sci Rep. 2016;6:38750. https://doi.org/10.1038/srep38750.
Kanda M, Shimizu D, Nomoto S, Takami H, Hibino S, Oya H, et al. Prognostic impact of expression and methylation status of DENN/MADD domain-containing protein 2D in gastric cancer. Gastric Cancer. 2015;18(2):288–96. https://doi.org/10.1007/s10120-014-0372-0.
Kanda M, Nomoto S, Oya H, Takami H, Hibino S, Hishida M, et al. Downregulation of DENND2D by promoter hypermethylation is associated with early recurrence of hepatocellular carcinoma. Int J Oncol. 2014;44(1):44–52. https://doi.org/10.3892/ijo.2013.2165.
Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25(10):1010–22.
Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013;38(1):23–38.
Guo W, Wang H, Li C. Signal pathways of melanoma and targeted therapy. Signal Transduct Target Ther. 2021;6(1):424.
Maner BS, Dupuis L, Su A, Jueng JJ, Harding TP, Meisenheimer J, et al. Overview of genetic signaling pathway interactions within cutaneous malignancies. J Cancer Metastasis Treat. 2020;6:37.
Sheng Y, Li F, Qin Z. TNF receptor 2 makes tumor necrosis factor a friend of tumors. Front Immunol. 2018;9:1170. https://doi.org/10.3389/fimmu.2018.01170.
Trentin L, Zambello R, Bulian P, Cerutti A, Enthammer C, Cassatella M, et al. Tumour-infiltrating lymphocytes bear the 75 kDa tumour necrosis factor receptor. Br J Cancer. 1995;71(2):240–5. https://doi.org/10.1038/bjc.1995.50.
Arnott CH, Scott KA, Moore RJ, Robinson SC, Thompson RG, Balkwill FR. Expression of both TNF-α receptor subtypes is essential for optimal skin tumour development. Oncogene. 2004;23(10):1902–10.
Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, et al. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66(16):8293–6.
Tao S, Park SL, de la Vega MR, Zhang DD, Wondrak GT. Systemic administration of the apocarotenoid bixin protects skin against solar UV-induced damage through activation of NRF2. Free Radical Biol Med. 2015;89:690–700.
Singh S, Vrishni S, Singh BK, Rahman I, Kakkar P. Nrf2-ARE stress response mechanism: a control point in oxidative stress-mediated dysfunctions and chronic inflammatory diseases. Free Radical Res. 2010;44(11):1267–88. https://doi.org/10.3109/10715762.2010.507670.
Hu R, Saw CL, Yu R, Kong AN. Regulation of NF-E2-related factor 2 signaling for cancer chemoprevention: antioxidant coupled with antiinflammatory. Antioxid Redox Signal. 2010;13(11):1679–98. https://doi.org/10.1089/ars.2010.3276.
Deshmukh P, Unni S, Krishnappa G, Padmanabhan B. The Keap1-Nrf2 pathway: promising therapeutic target to counteract ROS-mediated damage in cancers and neurodegenerative diseases. Biophys Rev. 2017;9(1):41–56.
Kansanen E, Kuosmanen SM, Leinonen H, Levonen AL. The Keap1-Nrf2 pathway: mechanisms of activation and dysregulation in cancer. Redox Biol. 2013;1:45–9. https://doi.org/10.1016/j.redox.2012.10.001.
Acknowledgements
This paper is dedicated to Professor Michael B. Sporn, the discoverer of CDDO series of triterpenoids, making great contribution to cancer chemoprevention research, who passed away recently. In addition, the authors are thankful to Lujing Wang in Dr. Kong’s laboratory for providing helpful insight into the study design, data analyses, and manuscript writing. The graphical abstract was created with BioRender.com.
Funding
This work is supported in part by institutional funds, R01 CA200129, from the National Cancer Institute and AT009152 from the National Center for Complementary and Integrative Health (NCCIH) to A-NTK.
Author information
Authors and Affiliations
Contributions
Participated in research design: H-CDK, A-NTK. Conducted experiments: H-CDK. Performed data analysis: H-CDK, RW, MZ, CW, DS. Wrote the manuscript: H-CDK, MSS. Reviewed the manuscript: A-NTK, NS, MSS, H-CDK.
Corresponding author
Ethics declarations
Conflict of Interest
NS is an inventor on patents dealing with chemical synthesis of triterpenoids and their application in cancer as well as inflammatory diseases. The rest of the authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kuo, HC.D., Wu, R., Sarwar, M.S. et al. DNA Methylome and Transcriptome Study of Triterpenoid CDDO in TPA-Mediated Skin Carcinogenesis Model. AAPS J 24, 115 (2022). https://doi.org/10.1208/s12248-022-00763-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-022-00763-5