Abstract
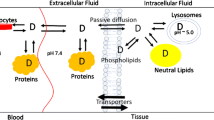
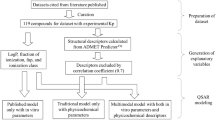
In physiologically based pharmacokinetic (PBPK) modelling, the large number of input parameters, limited amount of available data and the structural model complexity generally hinder simultaneous estimation of uncertain and/or unknown parameters. These parameters are generally subject to estimation. However, the approaches taken for parameter estimation vary widely. Global sensitivity analyses are proposed as a method to systematically determine the most influential parameters that can be subject to estimation. Herein, a global sensitivity analysis was conducted to identify the key drug and physiological parameters influencing drug disposition in PBPK models and to potentially reduce the PBPK model dimensionality. The impact of these parameters was evaluated on the tissue-to-unbound plasma partition coefficients (Kpus) predicted by the Rodgers and Rowland model using Latin hypercube sampling combined to partial rank correlation coefficients (PRCC). For most drug classes, PRCC showed that LogP and fraction unbound in plasma (fup) were generally the most influential parameters for Kpu predictions. For strong bases, blood:plasma partitioning was one of the most influential parameter. Uncertainty in tissue composition parameters had a large impact on Kpu and Vss predictions for all classes. Among tissue composition parameters, changes in Kpu outputs were especially attributed to changes in tissue acidic phospholipid concentrations and extracellular protein tissue:plasma ratio values. In conclusion, this work demonstrates that for parameter estimation involving PBPK models and dimensionality reduction purposes, less influential parameters might be assigned fixed values depending on the parameter space, while influential parameters could be subject to parameters estimation.




Similar content being viewed by others
References
Jones HM, Chen Y, Gibson C, Heimbach T, Parrott N, Peters SA, et al. Physiologically based pharmacokinetic modeling in drug discovery and development: a pharmaceutical industry perspective. Clin Pharmacol Ther. 2015;97(3):247–62. https://doi.org/10.1002/cpt.37.
Jamei M. Recent advances in development and application of physiologically-based pharmacokinetic (PBPK) models: a transition from academic curiosity to regulatory acceptance. Curr Pharmacol Rep. 2016;2:161–9. https://doi.org/10.1007/s40495-016-0059-9.
Luzon E, Blake K, Cole S, Nordmark A, Versantvoort C, Berglund EG. Physiologically based pharmacokinetic modeling in regulatory decision-making at the European Medicines Agency. Clin Pharmacol Ther. 2017;102:98-105. https://doi.org/10.1002/cpt.539.
Poulin P, Theil FP. A priori prediction of tissue:plasma partition coefficients of drugs to facilitate the use of physiologically-based pharmacokinetic models in drug discovery. J Pharm Sci. 2000;89(1):16–35. https://doi.org/10.1002/(sici)1520-6017(200001)89:1<16::aid-jps3>3.0.co;2-e.
Rodgers T, Leahy D, Rowland M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate-to-strong bases. J Pharm Sci. 2005;94(6):1259–76. https://doi.org/10.1002/jps.20322.
Berezhkovskiy LM. Volume of distribution at steady state for a linear pharmacokinetic system with peripheral elimination. J Pharm Sci. 2004;93(6):1628–40. https://doi.org/10.1002/jps.20073.
Schmitt W. General approach for the calculation of tissue to plasma partition coefficients. Toxicol in Vitro. 2008;22(2):457–67. https://doi.org/10.1016/j.tiv.2007.09.010.
Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci. 2006;95(6):1238–57. https://doi.org/10.1002/jps.20502.
Rodgers T, Rowland M. Mechanistic approaches to volume of distribution predictions: understanding the processes. Pharm Res. 2007;24(5):918–33. https://doi.org/10.1007/s11095-006-9210-3.
Graham H, Walker M, Jones O, Yates J, Galetin A, Aarons L. Comparison of in-vivo and in-silico methods used for prediction of tissue: plasma partition coefficients in rat. J Pharm Pharmacol. 2012;64(3):383–96. https://doi.org/10.1111/j.2042-7158.2011.01429.x.
Jones RD, Jones HM, Rowland M, Gibson CR, Yates JW, Chien JY, et al. PhRMA CPCDC initiative on predictive models of human pharmacokinetics, part 2: comparative assessment of prediction methods of human volume of distribution. J Pharm Sci. 2011;100(10):4074–89. https://doi.org/10.1002/jps.22553.
Woodruff TJ, Bois FY. Optimization issues in physiological toxicokinetic modeling: a case study with benzene. Toxicol Lett. 1993;69(2):181–96.
Yang J, Jamei M, Heydari A, Yeo KR, de la Torre R, Farre M, et al. Implications of mechanism-based inhibition of CYP2D6 for the pharmacokinetics and toxicity of MDMA. J Psychopharmacol. 2006;20(6):842–9. https://doi.org/10.1177/0269881106065907.
Peters SA. Identification of intestinal loss of a drug through physiologically based pharmacokinetic simulation of plasma concentration-time profiles. Clin Pharmacokinet. 2008;47(4):245–59. https://doi.org/10.2165/00003088-200847040-00003.
Xia B, Heimbach T, Gollen R, Nanavati C, He H. A simplified PBPK modeling approach for prediction of pharmacokinetics of four primarily renally excreted and CYP3A metabolized compounds during pregnancy. AAPS J. 2013;15(4):1012–24. https://doi.org/10.1208/s12248-013-9505-3.
Ke A, Barter Z, Rowland-Yeo K, Almond L. Towards a best practice approach in PBPK modeling: case example of developing a unified Efavirenz model accounting for induction of CYPs 3A4 and 2B6. CPT Pharmacometrics Syst Pharmacol. 2016;5(7):367–76. https://doi.org/10.1002/psp4.12088.
Budha NR, Ji T, Musib L, Eppler S, Dresser M, Chen Y, et al. Evaluation of cytochrome P450 3A4-mediated drug-drug interaction potential for cobimetinib using physiologically based pharmacokinetic modeling and simulation. Clin Pharmacokinet. 2016;55(11):1435–45. https://doi.org/10.1007/s40262-016-0412-5.
Tsamandouras N, Rostami-Hodjegan A, Aarons L. Combining the ‘bottom up’ and ‘top down’ approaches in pharmacokinetic modelling: fitting PBPK models to observed clinical data. Br J Clin Pharmacol. 2015;79(1):48–55. https://doi.org/10.1111/bcp.12234.
European Medicines Agency, Guideline on the qualification and reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation. 2018 [May 2019]; Available from: https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-reporting-physiologically-based-pharmacokinetic-pbpk-modelling-simulation_en.pdf.
U.S. Food and Drug Administration, Physiologically based pharmacokinetic analyses—format and content : guidance for industry. 2018 [May 2019]; Available from: https://www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/UCM531207.pdf.
Graham H. Predicting drug distribution in rat and human: University of Manchester; 2012.
Davies B, Morris T. Physiological parameters in laboratory animals and humans. Pharm Res. 1993;10(7):1093–5.
Rowland M, Tozer TN. Clinical pharmacokinetics: concepts and applications. 3rd ed. Baltimore: Lippincott Williams & Wilkins; 1995.
Kamboj S, Cheng JJ, Yu C. Deterministic vs. probabilistic analyses to identify sensitive parameters in dose assessment using RESRAD. Health Phys. 2005;88(5 Suppl):S104–9.
Hamby DM. A review of techniques for parameter sensitivity analysis of environmental models. Environ Monit Assess. 1994;32(2):135–54. https://doi.org/10.1007/BF00547132.
McKay MD, Beckman RJ, Conover WJ. Comparison of three methods for selecting values of input variables in the analysis of output from a computer code. Technometrics. 1979;21(2):239–45. https://doi.org/10.1080/00401706.1979.10489755.
Blower SM, Dowlatabadi H. Sensitivity and uncertainty analysis of complex-models of disease transmission—an Hiv model, as an example. Int Stat Rev. 1994;62(2):229–43. https://doi.org/10.2307/1403510.
Marino S, Hogue IB, Ray CJ, Kirschner DE. A methodology for performing global uncertainty and sensitivity analysis in systems biology. J Theor Biol. 2008;254(1):178–96. https://doi.org/10.1016/j.jtbi.2008.04.011.
Saltelli A, Chan K, Scott EM. Sensitivity analysis. Chichester. New York: Wiley; 2000.
R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria 2018.
Carnell R. lhs: Latin Hypercube Samples. R package version 1.0.1 ed: Comprehensive R Archive Network (CRAN); 2019.
Margolskee A, Darwich AS, Pepin X, Pathak SM, Bolger MB, Aarons L, et al. IMI - oral biopharmaceutics tools project—evaluation of bottom-up PBPK prediction success part 1: characterisation of the OrBiTo database of compounds. Eur J Pharm Sci. 2017;96:598–609. https://doi.org/10.1016/j.ejps.2016.09.027.
Poulin P, Jones HM, Jones RD, Yates JW, Gibson CR, Chien JY, et al. PhRMA CPCDC initiative on predictive models of human pharmacokinetics, part 1: goals, properties of the PhRMA dataset, and comparison with literature datasets. J Pharm Sci. 2011;100(10):4050–73. https://doi.org/10.1002/jps.22554.
Abdi H. The Bonferonni and Šidák corrections for multiple comparisons. Encyclopedia of measurement and statistics 2007;3.
Yamazaki K, Kanaoka M. Computational prediction of the plasma protein-binding percent of diverse pharmaceutical compounds. J Pharm Sci. 2004;93(6):1480–94. https://doi.org/10.1002/jps.20059.
Laznicek M, Laznickova A. The effect of lipophilicity on the protein binding and blood cell uptake of some acidic drugs. J Pharm Biomed Anal. 1995;13(7):823–8. https://doi.org/10.1016/0731-7085(95)01504-e.
Ghafourian T, Amin Z. QSAR models for the prediction of plasma protein binding. Bioimpacts. 2013;3(1):21–7. https://doi.org/10.5681/bi.2013.011.
Iman RL, Conover WJ. A distribution-free approach to inducing rank correlation among input variables. Commun Stat B Simul. 1982;11(3):311–34. https://doi.org/10.1080/03610918208812265.
Saltelli A. Global sensitivity analysis: the primer. Chichester, England. Hoboken: John Wiley; 2008.
Saltelli A, Ratto M, Tarantola S, Campolongo F, Commission E, Ispra JRC. Sensitivity analysis practices: strategies for model-based inference. Reliab Eng Syst Safe. 2006;91(10–11):1109–25. https://doi.org/10.1016/j.ress.2005.11.014.
Poulin P, Theil FP. Development of a novel method for predicting human volume of distribution at steady-state of basic drugs and comparative assessment with existing methods. J Pharm Sci. 2009;98(12):4941–61. https://doi.org/10.1002/jps.21759.
Poulin P, Jones RD, Jones HM, Gibson CR, Rowland M, Chien JY, et al. PHRMA CPCDC initiative on predictive models of human pharmacokinetics, part 5: prediction of plasma concentration-time profiles in human by using the physiologically-based pharmacokinetic modeling approach. J Pharm Sci. 2011;100(10):4127–57. https://doi.org/10.1002/jps.22550.
Simon G, Rouser G. Species variations in phospholipid class distribution of organs. II. Heart and skeletal muscle. Lipids. 1969;4(6):607–14.
Rouser G, Simon G, Kritchevsky G. Species variations in phospholipid class distribution of organs. I. Kidney, liver and spleen. Lipids. 1969;4(6):599–606.
Hof H, Simon RG. Phospholipid content of human and Guinea pig muscle: post-mortem changes and variations with muscle composition. Lipids. 1970;5(5):485–7.
Diagne A, Fauvel J, Record M, Chap H, Douste-Blazy L. Studies on ether phospholipids. II. Comparative composition of various tissues from human, rat and guinea pig. Biochim Biophys Acta. 1984;793(2):221–31.
Miller SP, Zirzow GC, Doppelt SH, Brady RO, Barton NW. Analysis of the lipids of normal and Gaucher bone marrow. J Lab Clin Med. 1996;127(4):353–8.
Ellmerer M, Schaupp L, Brunner GA, Sendlhofer G, Wutte A, Wach P, et al. Measurement of interstitial albumin in human skeletal muscle and adipose tissue by open-flow microperfusion. Am J Physiol Endocrinol Metab. 2000;278(2):E352–6. https://doi.org/10.1152/ajpendo.2000.278.2.E352.
Aitchison J, Shen SM. Logistic-normal distributions—some properties and uses. Biometrika. 1980;67(2):261–72. https://doi.org/10.2307/2335470.
Tsamandouras N, Wendling T, Rostami-Hodjegan A, Galetin A, Aarons L. Incorporation of stochastic variability in mechanistic population pharmacokinetic models: handling the physiological constraints using normal transformations. J Pharmacokinet Pharmacodyn. 2015;42(4):349–73. https://doi.org/10.1007/s10928-015-9418-0.
Berezhkovskiy LM. Determination of volume of distribution at steady state with complete consideration of the kinetics of protein and tissue binding in linear pharmacokinetics. J Pharm Sci. 2004;93(2):364–74. https://doi.org/10.1002/jps.10539.
Peyret T, Poulin P, Krishnan K. A unified algorithm for predicting partition coefficients for PBPK modeling of drugs and environmental chemicals. Toxicol Appl Pharmacol. 2010;249(3):197–207. https://doi.org/10.1016/j.taap.2010.09.010.
Poulin P, Haddad S. Advancing prediction of tissue distribution and volume of distribution of highly lipophilic compounds from a simplified tissue-composition-based model as a mechanistic animal alternative method. J Pharm Sci. 2012;101(6):2250–61. https://doi.org/10.1002/jps.23090.
Poulin P, Theil FP. Prediction of pharmacokinetics prior to in vivo studies. 1. Mechanism-based prediction of volume of distribution. J Pharm Sci. 2002;91(1):129–56.
Assmus F, Houston JB, Galetin A. Incorporation of lysosomal sequestration in the mechanistic model for prediction of tissue distribution of basic drugs. Eur J Pharm Sci. 2017;109:419–30. https://doi.org/10.1016/j.ejps.2017.08.014.
Holt K, Ye M, Nagar S, Korzekwa KR. Prediction of tissue - plasma partition coefficients using microsomal partitioning: incorporation into physiologically-based pharmacokinetic models and steady state volume of distribution predictions. Drug Metab Dispos. 2019;47(10):1050-1060. https://doi.org/10.1124/dmd.119.087973.
Davis AM, Webborn PJ, Salt DW. Robust assessment of statistical significance in the use of unbound/intrinsic pharmacokinetic parameters in quantitative structure-pharmacokinetic relationships with lipophilicity. Drug Metab Dispos. 2000;28(2):103–6.
Laruelle M, Slifstein M, Huang Y. Relationships between radiotracer properties and image quality in molecular imaging of the brain with positron emission tomography. Mol Imaging Biol. 2003;5(6):363–75.
Laznicek M, Kvetina J, Mazak J, Krch V. Plasma protein binding-lipophilicity relationships: interspecies comparison of some organic acids. J Pharm Pharmacol. 1987;39(2):79–83.
Lobell M, Sivarajah V. In silico prediction of aqueous solubility, human plasma protein binding and volume of distribution of compounds from calculated pKa and AlogP98 values. Mol Divers. 2003;7(1):69–87.
van de Waterbeemd H, Smith DA, Jones BC. Lipophilicity in PK design: methyl, ethyl, futile. J Comput Aided Mol Des. 2001;15(3):273–86.
Kratochwil NA, Huber W, Muller F, Kansy M, Gerber PR. Predicting plasma protein binding of drugs: a new approach. Biochem Pharmacol. 2002;64(9):1355–74.
Liu JZ, Yang L, Li Y, Pan DH, Hopfinger AJ. Constructing plasma protein binding model based on a combination of cluster analysis and 4D-fingerprint molecular similarity analyses. Bioorgan Med Chem. 2006;14(3):611–21. https://doi.org/10.1016/j.bmc.2005.08.035.
Saiakhov RD, Stefan LR, Klopman G. Multiple computer-automated structure evaluation model of the plasma protein binding affinity of diverse drugs. Perspect Drug Discov. 2000;19(1):133–55. https://doi.org/10.1023/A:1008723723679.
Watanabe R, Esaki T, Kawashima H, Natsume-Kitatani Y, Nagao C, Ohashi R, et al. Predicting fraction unbound in human plasma from chemical structure: improved accuracy in the low value ranges. Mol Pharm. 2018;15(11):5302–11. https://doi.org/10.1021/acs.molpharmaceut.8b00785.
Gleeson MP. Plasma protein binding affinity and its relationship to molecular structure: an in-silico analysis. J Med Chem. 2007;50(1):101–12. https://doi.org/10.1021/jm060981b.
Plante J, Werner S. JPlogP: an improved logP predictor trained using predicted data. J Cheminformatics. 2018;10:61. https://doi.org/10.1186/s13321-018-0316-5.
Doki K, Darwich AS, Achour B, Tornio A, Backman JT, Rostami-Hodjegan A. Implications of intercorrelation between hepatic CYP3A4-CYP2C8 enzymes for the evaluation of drug-drug interactions: a case study with repaglinide. Br J Clin Pharmacol. 2018;84(5):972–86. https://doi.org/10.1111/bcp.13533.
Melillo N, Darwich AS, Magni P, Rostami-Hodjegan A. Accounting for inter-correlation between enzyme abundance: a simulation study to assess implications on global sensitivity analysis within physiologically-based pharmacokinetics. J Pharmacokinet Pharmacodyn. 2019;46:137–54. https://doi.org/10.1007/s10928-019-09627-6.
Vozeh S, Schmidlin O, Taeschner W. Pharmacokinetic drug data. Clin Pharmacokinet. 1988;15(4):254–82. https://doi.org/10.2165/00003088-198815040-00005.
Kalvass JC, Phipps C, Jenkins GJ, Stuart P, Zhang X, Heinle L, et al. Mathematical and experimental validation of flux dialysis method: an improved approach to measure unbound fraction for compounds with high protein binding and other challenging properties. Drug Metab Dispos. 2018;46(4):458–69. https://doi.org/10.1124/dmd.117.078915.
Da Veiga S, Wahl F, Gamboa F. Local polynomial estimation for sensitivity analysis on models with correlated inputs. Technometrics. 2009;51(4):452–63. https://doi.org/10.1198/Tech.2009.08124.
Saltelli A, Tarantola S. On the relative importance of input factors in mathematical models: safety assessment for nuclear waste disposal. J Am Stat Assoc. 2002;97(459):702–9. https://doi.org/10.1198/016214502388618447.
Gray GM, Yardley HJ. Lipid compositions of cells isolated from pig, human, and rat epidermis. J Lipid Res. 1975;16(6):434–40.
Ruark CD, Hack CE, Robinson PJ, Mahle DA, Gearhart JM. Predicting passive and active tissue:plasma partition coefficients: interindividual and interspecies variability. J Pharm Sci. 2014;103(7):2189–98. https://doi.org/10.1002/jps.24011.
Kuksis A. Fatty acid composition of glycerolipids of animal tissues. In: Kuksis A, editor. Fatty acids and glycerides. Boston: Springer US; 1978. p. 381–442.
Ulloa JL, Stahl S, Yates J, Woodhouse N, Kenna JG, Jones HB, et al. Assessment of gadoxetate DCE-MRI as a biomarker of hepatobiliary transporter inhibition. NMR Biomed. 2013;26(10):1258–70. https://doi.org/10.1002/nbm.2946.
Bergstrom M, Grahnen A, Langstrom B. Positron emission tomography microdosing: a new concept with application in tracer and early clinical drug development. Eur J Clin Pharmacol. 2003;59(5–6):357–66. https://doi.org/10.1007/s00228-003-0643-x.
Chang YJ, Chang CH, Chang TJ, Yu CY, Chen LC, Jan ML, et al. Biodistribution, pharmacokinetics and microSPECT/CT imaging of 188Re-bMEDA-liposome in a C26 murine colon carcinoma solid tumor animal model. Anticancer Res. 2007;27(4B):2217–25.
Holt K, Nagar S, Korzekwa K. Methods to predict volume of distribution. Current Pharmacology Reports. 2019;5(5):391–9. https://doi.org/10.1007/s40495-019-00186-5.
Korzekwa K, Nagar S. On the nature of physiologically-based pharmacokinetic models—a priori or a posteriori? Mechanistic or empirical? Pharm Res. 2017;34(3):529–34. https://doi.org/10.1007/s11095-016-2089-8.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 12481 kb)
Rights and permissions
About this article
Cite this article
Yau, E., Olivares-Morales, A., Gertz, M. et al. Global Sensitivity Analysis of the Rodgers and Rowland Model for Prediction of Tissue: Plasma Partitioning Coefficients: Assessment of the Key Physiological and Physicochemical Factors That Determine Small-Molecule Tissue Distribution. AAPS J 22, 41 (2020). https://doi.org/10.1208/s12248-020-0418-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-020-0418-7