Abstract
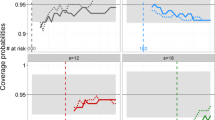
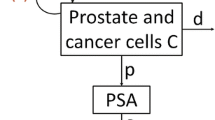
Given a joint model and its parameters, Bayesian individual dynamic prediction (IDP) of biomarkers and risk of event can be performed for new patients at different landmark times using observed biomarker values. The aim of the present study was to compare IDP, with uncertainty, using Stan 2.18, Monolix 2018R2 and NONMEM 7.4. Simulations of biomarker and survival were performed using a nonlinear joint model of prostate-specific antigen (PSA) kinetics and survival in metastatic prostate cancer. Several scenarios were evaluated, according to the strength of the association between PSA and survival. For various landmark times, a posteriori distribution of PSA kinetic individual parameters was estimated, given individual observations, with each software. Samples of individual parameters were drawn from the posterior distribution. Bias and imprecision of individual parameters as well as coverage of 95% credibility interval for PSA and risk of death were evaluated. All software performed equally well with small biases on individual parameters. Imprecision on individual parameters was comparable across software and showed marked improvements with increasing landmark time. In terms of coverage, results were also comparable and all software were able to well predict PSA kinetics and survival. As for computing time, Stan was faster than Monolix and NONMEM to obtain individual parameters. Stan 2.18, Monolix 2018R2 and NONMEM 7.4 are able to characterize IDP of biomarkers and risk of event in a nonlinear joint modelling framework with correct uncertainty and hence could be used in the context of individualized medicine.





Similar content being viewed by others
References
Tsiatis AA, Davidian M. Joint modeling of longitudinal and time-to-event data: An overview. Stat Sin. 2004;14(3):809–34.
Tsiatis AA, Degruttola V, Wulfsohn MS. Modeling the Relationship of Survival to Longitudinal Data Measured with Error. Applications to Survival and CD4 Counts in Patients with AIDS. J Am Stat Assoc. 1995;90(429):27–37.
Law NJ, Taylor JMG, Sandler H. The joint modeling of a longitudinal disease progression marker and the failure time process in the presence of cure. Biostat Oxf Engl. 2002;3(4):547–63.
Stanley CC, Kazembe LN, Buchwald AG, Mukaka M, Mathanga DP, Hudgens MG, et al. Joint modelling of time-to-clinical malaria and parasite count in a cohort in an endemic area. J Med Stat Inform 2019;7:1.
Barrett JK, Huille R, Parker R, Yano Y, Griswold M. Estimating the association between blood pressure variability and cardiovascular disease: An application using the ARIC Study. Stat Med. 2019;38(10):1855–68.
Rizopoulos D. Joint Models for Longitudinal and Time-to-Event Data : With Applications in R. Chapman and Hall/CRC; 2012.
Andrinopoulou E-R, Rizopoulos D, Geleijnse ML, Lesaffre E, Bogers AJJC, Takkenberg JJM. Dynamic prediction of outcome for patients with severe aortic stenosis: application of joint models for longitudinal and time-to-event data. BMC Cardiovasc Disord. 2015;15(1):28.
Sudell M, Kolamunnage-Dona R, Tudur-Smith C. Joint models for longitudinal and time-to-event data: a review of reporting quality with a view to meta-analysis. BMC Med Res Methodol 2016;16:168.
Desmée S, Mentré F, Veyrat-Follet C, Sébastien B, Guedj J. Using the SAEM algorithm for mechanistic joint models characterizing the relationship between nonlinear PSA kinetics and survival in prostate cancer patients. Biometrics. 2017;73(1):305–12.
Rizopoulos D. Dynamic Predictions and Prospective Accuracy in Joint Models for Longitudinal and Time-to-Event Data. Biometrics. 2011;67(3):819–29.
Tardivon C, Desmée S, Kerioui M, Bruno R, Wu B, Mentré F, et al. Association between tumor size kinetics and survival in urothelial carcinoma patients treated with atezolizumab: implication for patient’s follow-up. Clin Pharmacol Ther.
Desmée S, Mentré F, Veyrat-Follet C, Sébastien B, Guedj J. Nonlinear joint models for individual dynamic prediction of risk of death using Hamiltonian Monte Carlo: application to metastatic prostate cancer. BMC Med Res Methodol. 2017;17(1):105.
Stan Development Team. Stan Modeling Language User’s Guide and Reference Manual, Version 2.19.0. 2019. http://mc-stan.org/users/ documentation/index.html.
Lavielle M, Ribba B. Enhanced Method for Diagnosing Pharmacometric Models: Random Sampling from Conditional Distributions. Pharm Res. 2016;33(12):2979–88.
Plan EL, Maloney A, Mentré F, Karlsson MO, Bertrand J. Performance comparison of various maximum likelihood nonlinear mixed-effects estimation methods for dose-response models. AAPS J. 2012;14(3):420–32.
Björnsson MA, Friberg LE, Simonsson USH. Performance of Nonlinear Mixed Effects Models in the Presence of Informative Dropout. AAPS J. 2014;17(1):245–55.
Desmée S, Mentré F, Veyrat-Follet C, Guedj J. Nonlinear Mixed-effect Models for Prostate-specific Antigen Kinetics and Link with Survival in the Context of Metastatic Prostate Cancer: A Comparison by Simulation of Two-stage and Joint Approaches. AAPS J. 2015;17(3):691–9.
Lavielle M. Mixed Effects Models for the Population Approach: Models, Tasks, Methods and Tools. Chapman and Hall/CRC. 2014. (Chapman & Hall/CRC Biostatistics Series).
Neal RM, et al. MCMC using Hamiltonian dynamics. Handbook Markov Chain Monte Carlo. 2011;2:113–62.
Homan MD, Gelman A. The No-U-turn Sampler: Adaptively Setting Path Lengths in Hamiltonian Monte Carlo. J Mach Learn Res. 2014;15(1):1593–623.
Core Team R. R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing; 2018. https://www.R-project.org
Monolix 2018R1 User guide. Monolix 2017. http://monolix.lixoft.com/single-page/
Beal SL, Scheiner LB. NONMEM Users Guides. San Francisco: University of California; 1992.
Clopper CJ, Pearson ES. The Use of Confidence or Fiducial Limits Illustrated in the Case of the Binomial. Biometrika. 1934;26(4):404–13.
Chan PLS, Jacqmin P, Lavielle M, McFadyen L, Weatherley B. The use of the SAEM algorithm in MONOLIX software for estimation of population pharmacokinetic-pharmacodynamic-viral dynamics parameters of maraviroc in asymptomatic HIV subjects. J Pharmacokinet Pharmacodyn. 2011;38(1):41–61.
Touloumi G, Pocock SJ, Babiker AG, Darbyshire JH. Estimation and comparison of rates of change in longitudinal studies with informative drop-outs. Stat Med. 1999;18(10):1215–33.
Gastonguay MR, French JL, Heitjan DF, Rogers JA, Ahn JE, Ravva P. Missing Data in Model-Based Pharmacometric Applications: Points to Consider. J Clin Pharmacol. 2010;50(S9):63S–74S.
Blanche P, Proust-Lima C, Loubère L, Berr C, Dartigues J-F, Jacqmin-Gadda H. Quantifying and comparing dynamic predictive accuracy of joint models for longitudinal marker and time-to-event in presence of censoring and competing risks. Biometrics. 2015;71(1):102–13.
Graf E, Schmoor C, Sauerbrei W, Schumacher M. Assessment and comparison of prognostic classification schemes for survival data. Stat Med 1999.
Chapman MP, Tomlin CJ. Ordinary Differential Equations in Cancer Biology. bioRxiv. 2016;071134.
Claret L, Girard P, Hoff PM, Van Cutsem E, Zuideveld KP, Jorga K, et al. Model-Based Prediction of Phase III Overall Survival in Colorectal Cancer on the Basis of Phase II Tumor Dynamics. J Clin Oncol. 2009;27(25):4103–8.
Ribba B, Holford NH, Magni P, Trocóniz I, Gueorguieva I, Girard P, et al. A Review of Mixed-Effects Models of Tumor Growth and Effects of Anticancer Drug Treatment Used in Population Analysis. CPT Pharmacomet Syst Pharmacol. 2014;3(5):113.
Wilbaux M, Tod M, Bono JD, Lorente D, Mateo J, Freyer G, et al. A Joint Model for the Kinetics of CTC Count and PSA Concentration During Treatment in Metastatic Castration-Resistant Prostate Cancer. CPT Pharmacomet Syst Pharmacol. 2015;4(5):277–85.
Acknowledgments
The authors would like to acknowledge Dr. R. Bauer (ICON Development Solutions, Ellicott City, USA), for his valuable help in handling the NONMEM software. They also thank Hervé Le Nagard and Lionel de la Tribouille for the uses of CATIBioMed calculus facility.
Funding
François Riglet was supported by a grant from Sanofi, France.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that there is no conflict of interest regarding the publication of this article.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Riglet, F., Mentre, F., Veyrat-Follet, C. et al. Bayesian Individual Dynamic Predictions with Uncertainty of Longitudinal Biomarkers and Risks of Survival Events in a Joint Modelling Framework: a Comparison Between Stan, Monolix, and NONMEM. AAPS J 22, 50 (2020). https://doi.org/10.1208/s12248-019-0388-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-019-0388-9