Abstract
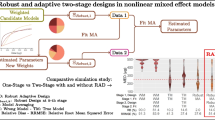
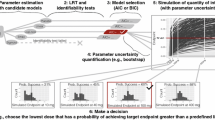
In drug development, pharmacometric approaches consist in identifying via a model selection (MS) process the model structure that best describes the data. However, making predictions using a selected model ignores model structure uncertainty, which could impair predictive performance. To overcome this drawback, model averaging (MA) takes into account the uncertainty across a set of candidate models by weighting them as a function of an information criterion. Our primary objective was to use clinical trial simulations (CTSs) to compare model selection (MS) with model averaging (MA) in dose finding clinical trials, based on the AIC information criterion. A secondary aim of this analysis was to challenge the use of AIC by comparing MA and MS using five different information criteria. CTSs were based on a nonlinear mixed effect model characterizing the time course of visual acuity in wet age-related macular degeneration patients. Predictive performances of the modeling approaches were evaluated using three performance criteria focused on the main objectives of a phase II clinical trial. In this framework, MA adequately described the data and showed better predictive performance than MS, increasing the likelihood of accurately characterizing the dose-response relationship and defining the minimum effective dose. Moreover, regardless of the modeling approach, AIC was associated with the best predictive performances.





Similar content being viewed by others
References
Cross J, Lee H, Westelinck A, Nelson J, Grudzinskas C, Peck C. Postmarketing drug dosage changes of 499 FDA-approved new molecular entities, 1980–1999. Pharmacoepidemiol Drug Saf. 2002;11(6):439–46.
Sacks LV, Shamsuddin HH, Yasinskaya YI, Bouri K, Lanthier ML, Sherman RE. Scientific and regulatory reasons for delay and denial of FDA approval of initial applications for new drugs, 2000–2012. JAMA. 2014;311(4):378–84.
Musuamba FT, Manolis E, Holford N, Cheung S, Friberg LE, Ogungbenro K, et al. Advanced methods for dose and regimen finding during drug development: summary of the EMA/EFPIA workshop on dose finding (London 4–5 December 2014). CPT Pharmacomet Syst Pharmacol. 2017;6(7):418–29.
Dose-response information to support drug registration (ICH Harmonized Tripartite Guideline), Page 2. 1994 [cited 2017 Jun 14]. Available from: http://www.ich.org/products/guidelines/efficacy/efficacy-single/article/dose-response-information-to-support-drug-registration.html
Bornkamp B, Bretz F, Dmitrienko A, Enas G, Gaydos B, Hsu C-H, et al. Innovative approaches for designing and analyzing adaptive dose-ranging trials. J Biopharm Stat. 2007;17(6):965–95.
Karlsson KE, Vong C, Bergstrand M, Jonsson EN, Karlsson MO. Comparisons of analysis methods for proof-of-concept trials. CPT Pharmacomet Syst Pharmacol. 2013;2(1):e23.
Bretz F, Pinheiro JC, Branson M. Combining multiple comparisons and modeling techniques in dose-response studies. Biometrics. 2005 Sep;61(3):738–48.
Pinheiro J, Bornkamp B, Glimm E, Bretz F. Model-based dose finding under model uncertainty using general parametric models. Stat Med. 2014;33(10):1646–61.
Buckland ST, Burnham KP, Augustin NH. Model selection: an integral part of inference. Biometrics. 1997;53(2):603–18.
Sébastien B, Hoffman D, Rigaux C, Pellissier F, Msihid J. Model averaging inconcentration–QT analyses. Pharm Stat. 2016;15(6):450–8.
Dosne AG, Bergstrand M, Karlsson MO, Renard D, Heimann G. Model averaging for robust assessment of QT prolongation by concentration-response analysis. Stat Med. 2017;36(24):3844–57.
Bertrand J, Comets E, Mentre F. Comparison of model-based tests and selection strategies to detect genetic polymorphisms influencing pharmacokinetic parameters. J Biopharm Stat. 2008;18(6):1084–102.
Bozdogan H. Model selection and Akaike’s information criterion (AIC): the general theory and its analytical extensions. Psychometrika. 1987;52(3):345–70.
Anderson DR, Burnham KP. Understanding information criteria for selection among capture-recapture or ring recovery models. Bird Study. 1999;46(sup1):S14–21.
Schorning K, Bornkamp B, Bretz F, Dette H. Model selection versus model averaging in dose finding studies. Stat Med. 2016;35(22):4021–40.
Bates DM. Nonlinear mixed effects models for longitudinal data. In: Wiley StatsRef: Statistics Reference Online [Internet]. John Wiley & Sons, Ltd; 2014. Available from: doi: https://doi.org/10.1002/9781118445112.stat05806/abstract
Holford N. Clinical pharmacology = disease progression + drug action. Br J Clin Pharmacol. 2015;79(1):18–27.
Buatois S, Retout S, Frey N, Ueckert S. Item response theory as an efficient tool to describe a heterogeneous clinical rating scale in de novo idiopathic Parkinson’s disease patients. Pharm Res. 2017 Oct 1;34(10):2109–18.
Aoki Y, Röshammar D, Hamrén B, Hooker AC. Model selection and averaging of nonlinear mixed-effect models for robust phase III dose selection. J Pharmacokinet Pharmacodyn 2017; 1–17.
Delattre M, Lavielle M, Poursat M-A. A note on BIC in mixed-effects models. Electron J Stat. 2014;8:456–75.
Claeskens G, Hjort NL. Model selection and model averaging. 1 edition. Cambridge; New York: Cambridge University Press; 2008. 332 p.
Rosenfeld PJ, Brown DM, Heier JS, Boyer DS, Kaiser PK, Chung CY, et al. Ranibizumab for neovascular age-related macular degeneration. N Engl J Med. 2006;355(14):1419–31.
Thomas N, Sweeney K, Somayaji V. Meta-analysis of clinical dose–response in a large drug development portfolio. Stat Biopharm Res. 2014;6(4):302–17.
Kullback S. Information theory and statistics. New edition ed. Mineola, N.Y: Dover Publications; 1997. p. 432.
MacKay DJC. Information theory, inference and learning algorithms. 1st ed. Cambridge, UK; New York: Cambridge University Press; 2003. p. 640.
Beal S, Sheiner LB, Boeckmann A, Bauer RJ. NONMEM user’s guides. (1989–2009). Ellicott City, MD USA: Icon Development Solutions; 2009.
Thomas N. Hypothesis testing and bayesian estimation using a sigmoid Emax model applied to sparse dose-response designs. J Biopharm Stat. 2006;16(5):657–77.
Dragalin V, Hsuan F, Padmanabhan SK. Adaptive designs for dose-finding studies based on sigmoid Emax model. J Biopharm Stat. 2007;17(6):1051–70.
Funding
This work was financed by a CIFRE agreement (Conventions Industrielles de Formation par la Recherche) and was conducted under the supervision of the ANRT (Association Nationale Recherche Technologie). The CIFRE agreement is a partnership between a public laboratory and a company, here the UMR 1137 and INSTITUT ROCHE, respectively.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 658 kb)
Rights and permissions
About this article
Cite this article
Buatois, S., Ueckert, S., Frey, N. et al. Comparison of Model Averaging and Model Selection in Dose Finding Trials Analyzed by Nonlinear Mixed Effect Models. AAPS J 20, 56 (2018). https://doi.org/10.1208/s12248-018-0205-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1208/s12248-018-0205-x