Abstract
Background
Cotton (Gossypium hirsutum L.) is one of the most significant fibre and cash crops and plays an important role in Indian industrial and agricultural economies. However, over the years quantity and quality have been hampered by the pest leafhopper. Leafhopper alone has been shown to cause yield losses of up to 40%. In this study, screening and evaluation were performed to identify and categorize 100 cotton genotypes along with 5 checks as resistant, moderately resistant, sensitive and highly sensitive to leafhoppers.
Results
A total of hundred genotypes were evaluated along with five checks for leafhopper resistance. Based on the screening results, a total of 19 genotypes were resistant to leafhoppers, which was on par with the findings of the check KC 3. The contents of total soluble sugar, total soluble protein, and total free amino acids were significantly positively correlated with the mean grade, whereas total phenols content and trichome density were significantly negatively correlated with the susceptibility grade. However, based on screening and biochemical analysis, the genotypes KC 2, JR-23, Samaru-26-T, D 4, TCH 1728, RS 253, and B-61-1862 exhibited high resistance to leafhopper.
Conclusion
According to the findings of this study, choosing genotypes with high total phenolics content together with high trichome density and low contents of total soluble sugar, total soluble protein, and free amino acids may aid in the development of resistant genotypes.
Similar content being viewed by others
Background
Cotton (Gossypium hirsutum L., (2n = 4x = 52)) is an allotetraploid species and one of the most significant fibre and cash crops. It plays an important role in India’s industrial and agricultural economies. It provides the textile industry with the primary raw material (cotton fiber). Cotton is one of the world's oldest and most ancient crops and is cultivated worldwide. India accounts for 26% of global production (Rajput et al. 2023). In India, both the quality and quantity of cotton have decreased over the years. The key cause of yield reduction was pest attack (Murugesan et al. 2010). Among all the pests, the major constraints were sap-sucking pests (Syed et al. 2003; Rizwan et al. 2021). However, among sucking pests, the most serious threat is posed by the leafhopper (Halder et al. 2016; Manivannan et al. 2021). The leafhopper attacks cotton during the growing season in all regions of the plant. During feeding, poison is injected into the leaves, causing aberrant alterations such as reddening and marginal chlorosis. When there are an enormous number of hoppers, stunted growth occurs, and the leaves bend downwards leading to severe boll losses, which in turn affect cotton yield. A 25%–40% yield reduction was registered due to leafhoppers (Ahmed et al. 2005). As a result, pest management approaches must be investigated. The widespread usage of pesticides in combating hoppers has led to the ability of these pests to withstand high pesticide dosages without dying (Rohini et al. 2011). This leads to the use of additional pesticides to achieve good control measures, resulting in increased pesticide pollution and disruption of the natural ecological balance (El-Sherbeni et al. 2019). Therefore, alternative approaches should be considered for the control of these sucking insects. Host plant resistance (HPR) offers a cost-effective and ecologically friendly way to reduce sucking insect pests. Therefore, the adoption of resistant cultivars is one of the best alternative strategies (Nawab et al. 2014; Salman et al. 2011) for combating leaf hoppers.
A variety of metabolic reactions contribute to host plant resistance to insect invasion. Certain chemicals function as feeding stimulants to stimulate insect feeding, whereas other chemicals interfere with the insects by imparting nonpreference, and antibiosis, disrupting the insects' metabolism to hinder their growth, development, and reproduction. It includes the biochemical properties of a plant, such as the production of phenolics, total sugars, total amino acids, and total proteins (Sandhi et al. 2017; Manivannan et al. 2021). Bhoge et al. (2019) investigated the biochemical HPR traits of resistance in cotton and discovered that the resistant varieties exhibited higher levels of phenolics in comparison to susceptible types and witnessed a negative correlation between total phenols and leafhoppers. Previous studies on HPR in cotton have mostly concentrated on morphological traits with fewer reports on biochemical traits together with a limited number of genotypes. In contrast to these investigations, we employed a far wider variety of genotypes encompassing the morphobiochemical HPR characteristics, from poisonous to pleasant to insects, including larger concentrations of sugars and soluble proteins.
In light of the above findings, the main aim of this study was to screen and evaluate cotton (Gossypium hirsutum L.) germplasm for resistance to leafhoppers.
Results
Screening of the germplasm
The germplasm was screened, and grades were assigned based on the symptoms displayed by the plants. The mean grade index was computed based on the infection symptoms. Among the total genotypes (Table S1), 19 entries were recorded with a scoring grade of 1 and were on par with a check (KC3), namely, Samaru-26-T, TCH 1728, JR-23, Pelimond Cleveland, 415\49-45F X LSS 3\63, D 4, PRS 12, SH 169, RS 253, TCH 1742, TCH 1569, KC 2, RU 4\4, TCH 1807, Paula, TCH 1811, TCH 1820, B-61-1862, and Alabar-637. Fifty-six entries were recorded with a scoring grade of 2, and twenty-three genotypes were recorded with a scoring grade of 3. Six genotypes were recorded with a scoring grade of 4, viz., G 4836, Acala-1577-D, MU 2, Coker, D 16, and RS 267.
Evaluation of resistance to leafhoppers
Morphological traits conferring resistance to leafhoppers
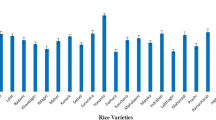
Trichome density evaluation revealed high variation in trichomes across the studied germplasm accessions (Table 1, Table S1, Fig. 1a). The trichome density varied from 125.50 to 420.50 cm−2. MU 2 had a lower trichome density (125.50 cm−2) but JR-23 had a high trichome density (420.50 cm−2). The genotypes viz., KC 2, JR-23, Samaru-26-T, D 4, SH 469, RS 253, and Paula showed trichome density on par with that of the check cultivar KC 3.
Biochemical parameters
The results revealed that the biochemical parameters, namely, total phenols content, total soluble sugar content, total free amino acids content, and total soluble protein content, exhibited extremely significant variability across the cotton genotypes (Table 1, Table S1, Table S2, Fig. 1b and Fig. 1c). The total soluble protein content varied from 20.99 to 53.41 mg·g−1, with KC 2 (20.99 mg·g−1) having the lowest total soluble protein. However, IC 263 SF (susceptible genotype) had the highest level (53.41 mg·g−1), which was greater than that of the check CO 17. The total soluble sugar content ranged from 3.68 to 7.70 mg·g−1, with genotype JR-23 (resistant) having the lowest level (3.68 mg·g−1) and G 4836 (highly susceptible) having the highest (7.70 mg·g−1). The total free amino acids content ranged from 17.07 to 24.44 mg·g−1, with KC 2 and BP-52 NC-62 containing the least amount (17.07 mg·g−1) and Tamcot SP 215 containing the most (24.44 mg·g−1). The total phenolic content varied from 2.38 to 6.65 µg·g−1, with MU 2 belonging to the susceptible class having the lowest content (2.38 µg·g−1) and the genotype RS 216 having the highest (6.65 µg·g−1).
Correlations between morphological and biochemical parameters and the mean grade index
The correlation analysis results are displayed in Table 2. The results revealed that total soluble protein content (r = 0.72**), total soluble sugar content (r = 0.61**), and total free amino acids content (r = 0.44**) were significantly positively correlated with the mean grade index. Total phenols content (r = −0.64**) and trichome density (r = −0.79**) were significantly negatively correlated with the mean grade index.
Discussion
Screening of the germplasm
The overall germplasms differed significantly from each other based on infection symptoms, which is in accordance with the findings of Bhoge et al. (2019). Based on the mean grade index, 19 entries were recorded with a grade of 1 and were on par with a check (KC 3), indicating that they were attacked less by the leafhopper. Hence, these plants are resistant, which may be due to the presence of high amounts of biochemical traits such as total phenols content (Neelima et al. 2010; Nishanth et al. 2016) and trichome density (Manivannan et al. 2017). Fifty-six accessions were found to be moderately resistant; twenty-three genotypes were found to be susceptible, and six genotypes were found to be highly susceptible, indicating that they have no inherent capability to resist the leafhopper throughout its growth stages. According to Bhoge et al. (2019) and Manivannan et al. (2021), susceptibility is due to low trichomes and high amounts of biochemical components such as total sugar, soluble protein, and total free amino acids.
Evaluation of resistance to leafhoppers
Morphological traits conferring resistance to leafhoppers
The evaluation of trichome density showed high variation in trichomes across the studied germplasm accessions which was also reported by Khalil et al. (2017), Pushpam and Ravikesavan (2019). Morphological studies revealed that trichome density was greater in plants with resistant/tolerant genotypes which confer resistance to leafhopper infestation. Manivannan et al. (2017) and Bhoge et al. (2019) reported that tolerant genotypes were found with a greater number of trichomes. As a result, leaf trichomes may be regarded as a key criterion for leafhopper resistance.
Biochemical parameters
Plant biochemical compounds have a significant impact on insect resistance. These compounds function as physiological inhibitors, feeding stimulants, or nutrient deficit inducers or by inhibiting insect development and metabolism. These findings were notable because of their antibiosis and nonpreference mechanisms for insect resistance (Rizwan et al. 2021). The results revealed that the biochemical parameters exhibited high variability across the cotton genotypes which is also supported by the findings of Bhoge et al. (2019). The estimation of contents of total soluble sugar, total free amino acids, and total soluble protein in the screened germplasms revealed that the susceptible genotypes had higher contents of these compounds in comparison to the resistant genotypes. These compounds act as feeding stimulants for insects (Praveen et al. 2013) which is in accordance with the findings of Pushpam and Ravikesavan (2019), and Murugesan et al. (2010) for total soluble protein, and Bhoge et al. (2019) for total soluble sugars. Manivannan et al. (2021) and Rizwan et al. (2021) also reported that leafhopper-susceptible varieties have a greater content of amino acids. Phenol serves as a vital line of defense, preventing insects from consuming it. The total phenolic content was found to be greater in the resistant genotypes. According to Constabel and Barbehenn (2008), phenolic compounds hamper the development and survival of insects. Similar findings were reported by Nishanth et al. (2016) and Pushpam and Ravikesavan (2019) who found that resistant genotypes have a greater phenol content than susceptible genotypes.
Correlations between morphological, and biochemical parameters and the mean grade index
The correlations among the studied parameters revealed that all the intercorrelations were significant. The results revealed a significantly positive correlation between total soluble protein content, total soluble sugar content and total free amino acids content and the mean grade index. This indicates that the sensitive genotypes have more of these biochemical compounds than the resistant genotypes. Pushpam and Ravikesavan (2019) and Rizwan et al. (2021) reported a significant positive correlation between these parameters and the index. However, total phenolics content and trichome density were significantly negatively correlated with the mean grade index, as they make plant sap less palatable to insects, which inhibits their capacity to consume plants, alters their metabolism, and reduces their ability to reproduce (War et al. 2012) which is consistent with the findings of Rizwan et al. (2021), Parveen et al. (2001), and Bhoge et al. (2019).
According to the observations stated above, leafhopper resistance seems to be governed by the plant’s intrinsic ability to produce a greater content of total phenol, more trichomes and leaf pubescence, and a lesser content of soluble sugars, free amino acids and soluble proteins.
Conclusion
The results of the present study demonstrated the genotypic injury indices and resistance categories of the different genotypes. Moreover, biochemical parameters and trichome density are important for determining leafhopper resistance and can be used as a benchmark for screening large populations for leafhopper resistance and narrowing down the progenies for field evaluation. Thus, they may assist the cotton grower in selecting varieties for their location by taking insect risk into account. It was also revealed that the selection and use of genotypes with higher phenols content and trichome density and a lower content of soluble sugar, amino acids, and soluble protein may be helpful for creating leafhopper-resistant genotypes. Thus, among the germplasm accessions, the genotypes that exhibit high resistance to leafhoppers namely KC 2, JR-23, Samaru-26-T, D 4, TCH 1728, RS 253, and B-61-1862 can be effectively used in an extensive hybridization program to increase the levels and broaden the sources of resistance against targeted insect pests.
Methods
This research study was conducted at the Department of Cotton, Tamil Nadu Agricultural University (TNAU), Coimbatore, India (2021–2022). The genotypes were raised in Augmented Block Design I (Federer and Raghavarao 1975), which consisted of a total of 100 germplasms along with 5 checks (KC 3, MCU 5, SVPR 6, CO 14, and CO 17) with a spacing of 90 cm (row to row) × 45 cm (plant to plant). The checks were replicated for every 10 rows of germplasm. During the whole trial, no pesticide was used to allow the cotton leafhopper to proliferate. All recommended agronomic techniques were used to increase crop output over the whole cropping period.
Screening of the germplasm
One hundred germplasm accessions along with five checks were screened under field conditions. Five randomly selected plants from each genotype were tagged. Three leaves (top, middle, bottom) were taken from the selected plants. The tagged plants from each entry were scored and graded on the 30th, 45th, 60th, and 75th day after sowing, based on the infection symptoms (Table 3) and the mean grade score of each entry was calculated. Based on the grade score, the entries were categorized into four groups: resistant (Grade 1), moderately resistant (Grade 2), susceptible (Grade 3), and highly susceptible (Grade 4) as per Sikka et al. (1966) with slight modifications.
Evaluation of resistance to leafhoppers
Morphological traits conferring resistance to leafhoppers
Fresh leaf samples were taken 45 days after sowing. Estimation was performed on the third fully grown leaf. Leaf samples were cut into 1 cm leaf pieces and boiled in 20 mL of water in small glass vials for 15 min before being removed. The leaf pieces were then heated in 20 mL of 96% (volume fraction) ethyl alcohol for approximately 20 min at 80 °C. After boiling, the alcohol was removed and the process was repeated with fresh alcohol until the chlorophyll content of the leaf fragments was completely removed. After removing the alcohol from the boiling tube, 90% (mass fraction) lactic acid was added to the vials, which were then sealed and heated at 85 °C for 35–45 min until the leaf segments were cleared. After the heating steps were completed, the vials were allowed to cool before the leaf fragments were extracted and placed on a glass slide with a pinching of lactic acid to count the trichomes. Trichomes in a one-centimeter area were counted using a Leica stereo zoom binocular microscope. The trichome density was expressed as trichomes per cm2 of leaf area.
Estimation of biochemical host plant-related traits
Total soluble proteins, total soluble sugars, free amino acids, and total phenols were estimated in the leaves of the cotton accessions. Five healthy plants from each genotype were selected 50 days after sowing. The third and the fourth opened fresh leaves from the top of each plant were used for biochemical analysis at the Department of Biochemistry, TNAU, Coimbatore, India. The biochemical components of the cotton cultures were noted during the experiment. This was achieved by adhering to the guidelines provided below.
a. Estimation of total soluble protein content
The estimation of total soluble protein was carried out using the technique given by Lowry et al. (1951). A 500 mg leaf sample was taken from each of the 100 genotypes, along with 5 checks. The samples were then centrifuged and extracted with the buffer. The sample (0.2 mL) extract was pipetted into a test tube. Working standard solutions of 0.2, 0.4, 0.6, 0.8, and 1.0 mL were pipetted into a series of test tubes. The volume of each tube was adjusted to 1 mL with distilled water. A tube with 1 mL of water served as a blank. Five milliliters of an alkaline copper solution were added to each tube, which was thoroughly mixed before being incubated for 10 min at room temperature. Then, the tubes were filled with 0.5 mL of Folin-Ciocalteu reagent, mixed immediately, and incubated at room temperature in the dark for 30 min. Then, the absorbance at 660 nm was measured against a reagent blank. A standard graph was generated, after which the total amount of protein in the sample was determined and expressed as mg·g−1.
b. Estimation of total soluble sugars
The estimation of total soluble sugars was performed using the standard technique given by Dubois et al. (1951). Leaf samples (50 mg) were extracted in a 2 mL solution of 95% (mass fraction) methanol. Sample extract, 5% (mass fraction) phenol, and 96% (mass fraction) sulfuric acid were used to create the assay solution. Leaf extract (1 mL), phenol (0.5 mL), and sulfuric acid (2.5 mL) were added to glass tubes. The tubes were carefully shaken, then placed in a water bath (26–30 °C) for 20 min. The absorbance of the colored solution was subsequently measured at 490 nm. Various concentrations of glucose (0.1–3.0 mg with 0.5 mg intervals and 3–6 mg with 1 mg intervals) were added to 4 mL of distilled water. The tubes were then placed in a water bath (26–30 °C) for 20 min. The absorbance was measured at 490 nm. A standard curve was plotted by plotting glucose concentration (x-axis) against the absorbance at 490 nm (y-axis).
c. Free amino acid estimation
The total free amino acids were assessed using the ninhydrin assay as described by Moore and Stein (1948). Leaf samples (500 mg) were taken from each of the 100 genotypes, along with 5 checks and ground with 10 mL of 80% (mass fraction) ethanol. The contents were centrifuged for 5 min at 10 000 r·min−1 after which the supernatant was collected. Working standard solutions of 0.2, 0.4, 0.6, 0.8, and 1.0 mL were pipetted into a series of test tubes. A total of 0.5 mL of sample extract was removed from the test tube. The test tubes were filled with 1 mL of ninhydrin reagent and thoroughly mixed. Distilled water was added to bring the volume up to 1 mL. Four milliliters of ninhydrin-citrate-glycerol were added and the mixture was thoroughly mixed. Following a 15-min boil, the tubes were allowed to cool under running water, and the absorbance of the purple color was measured at 570 nm (green filter) against a reagent blank. Using a standard curve made from leucine, which was obtained by pipetting out 0.1–1.0 mL (10–100 mg range) of the working standard solution, the total amount of free amino acids was determined and expressed as mg·g−1.
d. Estimation of total phenol content
The total phenolic content was determined using the Folin-Ciocalteu reagent, as described by Bray and Thorpe (1954). A 500 mg leaf sample was taken from each of the 100 genotypes, along with 5 checks and ground with 80% (mass fraction) ethanol. After incubating for 20 min, the homogenate was centrifuged at 10 000 r·min−1 after which the supernatant was collected. The residue was extracted five times in 80% ethanol, centrifuged, and then pooled with the supernatant, which was then evaporated to dryness. The sample was dissolved in distilled water to a final volume of 5 mL. The various aliquots (0.2 to 2 mL) were pipetted into test tubes, and the volume in each test tube was increased to 6 mL with distilled water. Folin-Ciocalteu reagent 0.5 mL was added. After 3 min, 2 mL of 20% (mass fraction) NaClO3 solution was added. The solution was combined thoroughly. After the tubes were cooled and submerged in boiling water for precisely one minute, the absorbance at 650 nm was determined against a reagent blank. A standard curve was developed using various concentrations of catechol. The concentration of phenols present in the test sample was determined and is expressed as μg·g−1.
Statistical analysis
The data collected were examined separately, an analysis of variance was applied, and correlation matrix was generated using IBM SPSS version 26. The data provided in the tables are the mean values.
Availability of data and materials
All the data generated or analyzed during this study are included in this published article.
References
Ahmed G, Arif MJ, Sanpal MRJ. Population fluctuation of Jassid, Amrasca devastans (Dist.) in cotton through morpho-physical plant traits. Caderno de Perquisa Ser Bio, Santa Cruz do sul. 2005;17(1):79.
Bhoge RS, Hole UB, Gangurde SM, et al. Biochemical and morphological factors responsible for jassid resistance in cotton. Int J Chem Stud. 2019;7(4):166–9.
Bray HG, Thorpe W. Analysis of phenolic compounds of interest in metabolism. Methods Biochem Anal. 1954;1:27–52. https://doi.org/10.1002/9780470110171.ch2.
Constabel CP, Barbehenn R. Defensive roles of polyphenol oxidase in plants. In: Schaller A, editor. Induced plant resistance to herbivory. Dordrecht: Springer Netherlands; 2008. p. 253–70. https://doi.org/10.1007/978-1-4020-8182-8_12.
Dubois M, Gilles KA, Hamilton JK, et al. A colorimetric method for the determination of sugars. Nat. 1951;168(4265):167. https://doi.org/10.1038/168167a0.
El-Sherbeni AEHED, Khaleid MS, AbdAllah SAEA, et al. Effect of some insecticides alone and in combination with salicylic acid against aphid, Aphis gossypii, and white fly Bemisia tabaci on cotton field. Bull Natl Res Cent. 2019;43(1):57. https://doi.org/10.1186/s42269-019-0103-0.
Federer WT, Raghavarao D. On augmented designs. Biometrics. 1975;31(1):29–35. https://doi.org/10.2307/2529707.
Halder J, Sanwal S, Deb D, et al. Mechanisms of physical and biochemical basis of resistance against leaf-hopper (Amrasca biguttula biguttula) in different okra (Abelmoschus esculentus) genotypes. Indian Agr Sci. 2016;86(4):481–4. https://doi.org/10.56093/ijas.v86i4.57457.
Khalil H, Raza ABM, Afzal M, et al. Effects of plant morphology on the incidence of sucking insect pests complex in few genotypes of cotton. J Saudi Soc Agric Sci. 2017;16(4):344–9. https://doi.org/10.1016/j.jssas.2015.11.003.
Lowry OH, Rosebrough NJ, Farr AL, et al. Protein measurement with Folin phenol reagent. J Biol Chem. 1951;193(1):265–75. https://doi.org/10.1016/s0021-9258(19)52451-6.
Manivannan A, Sridhar RP, Karthikeyan S, et al. Screening of cotton genotypes against leafhopper, Amrasca biguttula biguttula (Ishida) (Homoptera: Cicadellidae). J Entomol Zool Stud. 2017;5:1305–10.
Manivannan A, Kanjana D, Dharajothi B, et al. Evaluation of resistance in cotton genotypes against leafhoppers Amrasca biguttula biguttula (Ishida), (Homoptera: Cicadellidae). Int J Trop Insect Sci. 2021;41(2):2409–20. https://doi.org/10.1007/s42690-020-00416-0.
Moore S, Stein WH. Photometric ninhydrin method for use in chromatography of amino acids. J Biol Chem. 1948;176:367–88. https://doi.org/10.1016/s0021-9258(18)51034-6.
Murugesan N, Kavitha A. Host plant resistance in cotton accessions to the leaf hopper, Amrasca devastans I (Distant). J Biopestic. 2010;3(3):526–33.
Nawab NN, Mehmood A, Jeelani G, et al. Inheritance of okra leaf type, gossypol glands and trichomes in cotton. J Anim Plant Sci. 2014;24:526–33.
Neelima S, Rao Prasada GMV, et al. Reaction of cotton genotypes to leafhopper, Amrasca devastans. Indian J Plant Prot. 2010;38(2):147–51.
Nishanth GK, Harijan Y, Katageri IS. Screening for sucking pests (thrips and jassid) resistance/tolerance in cotton germplasm lines (Gossypium hirsutum L.). Bioscan. 2016;11(1):85–9.
Perveen SS, Qaisrani TM, Amin S, et al. Biochemical basis of insect resistance in cotton. J Biol Sci. 2001;1(6):496–500.
Praveen H, Ugalat J, Singh H. Biochemical changes during crop growth period of resistance and susceptible varieties of maize against stem borer. Environ Ecol. 2013;31(4):1621–6.
Pushpam R, Ravikesavan R. Morphological and biochemical basis of resistance to Jassid (Amrasca devastans) in cotton Gossypium hirsutum L. J Cot Res Dev. 2019;33(1):116–22.
Rajput A, Raghuwanshi PS, Chaturvedi P. Dynamics of area, production and productivity of cotton crop in India. Curr Agr Res J. 2023;11(2):CARJ11.2.12. https://doi.org/10.12944/carj.11.2.18.
Rizwan M, Abro S, Asif MU, et al. Evaluation of cotton germplasm for morphological and biochemical host plant resistance traits against sucking insect pest’s complex. J Cot Res. 2021;4:18. https://doi.org/10.1186/s42397-021-00093-5.
Rohini A, Prasad NVVSD, Chalam MSV, et al. Identification of suitable resistant cotton genotypes against sucking pests. J Entomol Res. 2021;35(3):197–202.
Salman M, Masood A, Arif MJ, et al. The resistance levels of different cotton varieties against sucking insect pests complex in Pakistan. Pak J Agr: Agr Engin Vet Sci. 2011;27(2):168–75.
Sandhi RK, Sidhu SK, Sharma A, et al. Morphological and biochemical basis of resistance in okra to cotton jassid, Amrasca biguttula (Ishida). Phytoparasitica. 2017;45:381–94. https://doi.org/10.1007/s12600-017-0589-7.
Sikka SM, Sahni VM, Butani DK. Studies on jassid resistance in relation to hairiness of cotton leaves. Euphytica. 1966;15(3):383–8. https://doi.org/10.1007/bf00022184.
Syed TS, Abro GH, Khuhro RD, Dhauroo MH. Relative resistance of cotton varieties against sucking pests. Pak J Bio Sci. 2003;6(14):1232–3. https://doi.org/10.3923/pjbs.2003.1232.1233.
War AR, Paulraj MG, Ahmad T, et al. Mechanisms of plant defense against insect herbivores. Plant Signaling Behav. 2012;7(10):1306–20. https://doi.org/10.4161/psb.21663.
Acknowledgements
The authors would like to thank the Department of Cotton and the Department of Biochemistry at Tamil Nadu Agricultural University, Coimbatore for providing the essential resources for this research.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Premalatha N conceived and designed the experiment. Mawblei C executed the experiment, analyzed the data, and drafted the paper. Manivannan A revised the manuscript. Senguttuvan K reviewed and formally analyzed the study. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent for publication
Not applicable.
Competing interest
The authors declare that they have no competing interests.
Supplementary Information
42397_2024_182_MOESM1_ESM.zip
Additional file 1: Table S1. Biochemical parameters and hopper injury status of the different genotypes. Table S2. Multiple comparisons within resistant grades.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Mawblei, C., Premalatha, N., Manivannan, A. et al. Characterization and screening of cotton (Gossypium hirsutum L.) germplasm for leafhopper (Amrasca biguttula biguttula (Ishida)) resistance. J Cotton Res 7, 19 (2024). https://doi.org/10.1186/s42397-024-00182-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42397-024-00182-1