Abstract
The inflammasome, typically consisting of a Nod-like receptor, apoptosis-associated speck-like protein, and pro-caspase-1, has recently been identified as a huge intracellular complex, which plays a crucial role in interleukin-1 maturation or specific physiological functions. Two Nod-like receptors, such as nucleotide-binding oligomerization domains-containing protein (Nod)1 and Nod2, interact with the receptor-interacting protein serine-threonine kinase (RIPK)2 accompanied by Iκ-B kinase (IKK) complexes to construct the nodosome, leading to nuclear factor (NF)-κB activation. The aberrant activation of inflammasomes or nodosomes causes autoinflammatory diseases. Therefore, inflammasomes may be attractive targets to treat autoinflammatory diseases. Our aim is to develop reconstituted inflammasomes in a cell-free system to discover specific molecular-target drugs and elucidate the molecular pathogenesis of autoinflammatory diseases. In this review, we describe reconstituted inflammasomes in a cell-free system.
Similar content being viewed by others
Background
Inflammasomes have recently been identified as expanding intracellular complexes that play important roles not only in innate immunity but also in maintaining specific physiological functions [1,2,3]. The aberrant activation of inflammasomes is thought to be linked to various diseases, including inflammatory diseases, degenerative diseases, and tumors [4, 5]. Therefore, inflammasomes may be attractive targets to treat these diseases. Autoinflammatory diseases are known to be caused by genetic mutations of inflammasome components [6,7,8,9]. Thus, we aim to develop reconstituted inflammasomes in a cell-free system in order to identify specific molecular-target drugs and elucidate the molecular pathogenesis of autoinflammatory diseases. In this review, we briefly describe the functions of several inflammasomes and related diseases, and reconstituted inflammasomes in a cell-free system.
General functions of inflammasomes and related diseases
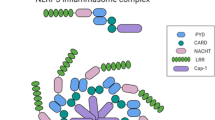
The inflammasomes have been known as interleukin (IL)-1β processing platforms [10, 11]. There are several well-characterized inflammasomes: NACHT, LRR (NLR), and PYD domain-containing protein (NLRP)1 inflammasome [11], NLRP3 inflammasome [12], absent in melanoma (AIM)2 inflammasome [13,14,15,16], NLR and CARD domain-containing protein (NLRC)4 inflammasome [17], and pyrin inflammasome [18]. The inflammasome typically consists of an intracellular pathogen pattern-recognition receptor, an adaptor protein apoptosis-associated speck-like protein containing a caspase recruitment domain (ASC), and pro-caspase-1.
The NLRP1 inflammasome was the first described inflammasome to be described [11]. It has been reported to be activated by muramyl dipeptide (MDP), anthrax lethal toxins, and related to neuronal diseases [19].
The NLRP3 inflammasome is a prototype inflammasome, activated by various pathogen-associated molecular pattern molecules (PAMPs) and damage-associated molecular pattern molecules (DAMPs) [20]. NLRP3-activated PAMPs have been reported to include bacterium-derived pore-forming toxins, lethal toxins, flagellin/rod proteins, MDP, RNA, DNA, virus-derived RNA, M2 protein, fungus-derived β-glucans, hypha mannan, zymosan, and protozoon-derived hemozoin [21]. NLRP3-activated DAMPs include self-derived ATP, cholesterol crystals, monosodium urate (MSU) crystals, calcium pyrophosphate dihydrate (CPPD) crystals, glucose, β-amyloid, hyaluronic acid, and environment-derived alum, asbestos, silica, alloy particles, UV radiation, and skin irritants [21].
A single amino acid mutation in NLRP3 results in enhanced inflammasome activation, termed cryopyrin-associated periodic syndrome (CAPS), including familial cold autoinflammatory syndrome (FCAS), Muckle–Wells syndrome (MWS), and neonatal-onset multisystem inflammatory disease (NOMID)/chronic infantile neurologic, cutaneous, and arthritis (CINCA) syndrome, which leads to greater IL-1β secretion without DAMPs or PAMPs [22,23,24,25,26,27].
The AIM2 inflammasome consists of AIM2, ASC, and pro-caspase-1. AIM2 was originally identified as an interferon-gamma inducible gene product consisting of an N-terminal pyrin domain (PYD) and C-terminal hematopoietic interferon-inducible nuclear proteins with a 200-amino acid repeat (HIN-200) domain. AIM2 is differentially expressed following the suppression of the tumorigenic phenotype in a malignant melanoma cell line [28], and it subsequently acts as a sensor for cytoplasmic DNA, which forms an inflammasome with the ligand and ASC to activate caspase-1 [13,14,15,16]. The inappropriate recognition of cytoplasmic self-DNA by AIM2 contributes to the development of psoriasis, dermatitis, arthritis, and other autoimmune and inflammatory diseases [29].
The NLRC4 inflammasome consists of NLRC4, ASC, and pro-caspase-1. Since the protein-binding motif of NLRC4 is CARD instead of the PYD, NLRC4 interacts with ASC as well as pro-caspase-1 through their CARD. NLRC4 constitutes an inflammasome, which is required for the recognition of bacterial flagellin [30, 31]. Several mutations in the nucleotide-binding domain of NLRC4 cause autoinflammatory diseases, early-onset recurrent fever flares, and macrophage activation syndrome (MAS) [32,33,34].
Pyrin has been identified as a causative gene of the MEFV product of familial Mediterranean fever (FMF), an autosomal recessive inherited autoinflammatory syndrome [35]. Pyrin not only regulates several inflammasomes [36,37,38] but also constructs an inflammasome with ASC and pro-caspase-1, upon recognizing some pathogens [18, 39, 40]. Thus, FMF patients with some pyrin mutations are thought to show autosomal dominant inheritance [41,42,43].
General functions of the nodosomes and related diseases
Nod1 and Nod2, both of which are involved in host recognition of small molecules, activate NF-κB in response to sensing the component of peptidoglycan [44,45,46,47]. NF-κB activation in Nod1 and Nod2 depends on RIPK2 and IKK machinery [48]. The core ligand structure of Nod2 is N-Acetyl muramyl-l-alanyl-d-isoglutamine hydrate, also known as MDP, of which the structure is common in bacteria. The ligand for Nod1 is a dipeptide designated as d-glutamyl-meso-diaminopimelic acid (iE-DAP), with the structure being derived from a subgroup of bacteria [44,45,46,47].
Functional activation by genetic mutations of Nod2 is associated with autoinflammatory diseases, Blau syndrome (BS), and early-onset sarcoidosis (EOS), which are characteristics of systemic granulomatous diseases [49]. However, genetic and functional defects of Nod2 are associated with susceptibility to Crohn’s disease, an inflammatory bowel disease. There is no known Nod1-related autoinflammatory disease, but associations between SNPs in NOD1 and several immune-related diseases, such as inflammatory bowel disease, atopic eczema, asthma, and rheumatoid arthritis have been reported [50,51,52,53].
Wheat germ cell-free protein synthesis for inflammasomes
To construct reconstituted inflammasomes in a cell-free system, we employed the wheat germ cell-free protein synthesis system rather than Escherichia coli expression system [54]. When we identified ASC a central adaptor protein of inflammasomes, ASC was discovered in the Triton X-100-insoluble fraction of promyelocytic leukemia cell line HL-60 cells [55], and it was difficult to synthesize recombinant NLRP3 protein using E. coli expression due to its solubility. On the other hand, the wheat germ cell-free protein synthesis has numerous advantages, such as low cost, ease of availability in large amounts, low endogenous incorporation, and the capacity to synthesize high-molecular-weight proteins [54]. In addition, it is suitable for the expression of eukaryotic proteins because it is eukaryotic system [55].
Reconstituted AIM2 inflammasome in a cell-free system
First, we describe an AIM2 inflammasome in a cell-free system as a prototype [56] because the AIM2 inflammasome has been well-characterized, and its ligand was reported to be present in poly-deoxyadenylic-deoxythymidylic acid, poly(dA:dT). The direct interaction between AIM2 and poly(dA:dT) was elucidated using the amplified luminescent proximity homogeneous assay (Alpha) [14]. In addition, activation of the AIM2 inflammasome has been reported to be related to various diseases [57,58,59,60,61,62], and it is thought to be an attractive drug target for diseases.
Our reconstituted AIM2 inflammasome basically consists of AIM2 and ASC, and it is considered sufficient for drug and ligand discovery as it assembles without pro-caspase-1 or any other components [56].
To synthesize the AIM2 inflammasome, PCR products for AIM2 and ASC were inserted into a Gateway™ pDONRTM221 Vector (pDONR221) (Life Technologies, Carlsbad, CA, USA) using Gateway™ BP Clonase™ II Enzyme mix (Life Technologies, Carlsbad, CA, USA) to generate entry clones. The AIM2 entry clone pDONR221-AIM2 was inserted into pEU-E01-GW-bls-STOP for cell-free protein expression. The ASC entry clones pDONR221-ASC and pDONR221-ASC-PYD were inserted into pEU-E01-FLAG-GW-STOP using the Gateway™ LR Clonase™ II Enzyme mix (Life Technologies, Carlsbad, CA, USA). The constructed plasmids were used to synthesize specific proteins with the WEPRO1240 Expression Kit (Cell-Free, Inc., Matsuyama, Japan) [56].
In our AIM2 inflammasome, proximity between AIM2 and ASC is detected by the Alpha using the combination of protein-A-conjugated Alpha acceptor beads for FLAG-tagged proteins and streptavidin-conjugated Alpha donor beads for biotinylated proteins (Fig. 1).
The AIM2 inflammasome in a cell-free system assembles with its previously reported ligand poly(dA:dT), and the interaction between AIM2 and ASC was disrupted by anti-human ASC mAb, and previously reported inhibitors CRID3 and glycyrrhizin. Thus, our reconstituted AIM2 inflammasome in a cell-free system is useful for investigating novel ligands and drug discovery [56].
Reconstituted NLRP3 inflammasome in a cell-free system
When AIM2 is replaced by NLRP3, we can easily develop the NLRP3 inflammasome in a cell-free system. There are so many mutations in NLRP3 that causes of autoinflammatory diseases including CAPS, and NLRP3 involve various inflammasomopathies. Thus, the reconstituted NLRP3 inflammasome in a cell-free system will be a useful tool for investigating inflammasomopathies and drug discovery. In this context, we are going to develop reconstituted NLRP3 inflammasome in a cell-free system.
Reconstituted Nod2 nodosome in a cell-free system
The autoinflammatory disease Blau syndrome (BS)/early-onset sarcoidosis (EOS) is caused by a point mutation of Nod2 [49]. Therefore, the Nod2 nodosome may be an attractive drug target for the treatment of BS/EOS. We aimed to develop a reconstituted protein–protein interaction assay system between wild-type Nod2 and the BS/EOS-associated mutants of Nod2 and RIPK2 in a cell-free system, called the reconstituted Nod2 nodosome in a cell-free system [63].
The plasmids vector pDONR221-Nod2 and BS/EOS-associated mutants, pDONR221-Nod2-R334W and pDONR221-Nod2-N670K, were constructed. pDONR221-RIPK2 and pDONR221-RIPK2-CARD were also constructed. Then, the proteins Nod2-WT-Btn, Nod2-R334W-Btn, Nod2-N670K-Btn, FLAG-RIPK2 and FLAG-RIPK2-CARD were synthesized using the wheat germ cell-free system in the same way as AIM2.
In our Nod2 nodosome, proximity between Nod2 and RIPK2 is basically detected by Alpha using the combination of protein-A-conjugated Alpha acceptor beads for FLAG-tagged proteins and streptavidin-conjugated Alpha donor beads for biotinylated proteins. The Nod2 nodosome in a cell-free system assembles with its previously reported ligand MDP. The Nod2 nodosomes with BS/EOS-associated mutations Nod2-R334W and Nod2-N670K were more sensitive to MDP than Nod2-WT. Therefore, we think that our Nod2 nodosome in a cell-free system can be a useful tool for investigating the pathogenesis of BS/EOS and drug discovery [63].
How does the reconstituted inflammasome in a cell-free system work?
We show representative data that suggest the mechanism of how this system works (Fig. 2). We synthesized two truncated forms of AIM2, AIM2-PYD-FLAG and AIM-HIN-200-Biotin, using a wheat germ cell-free synthesis system (Fig. 2a). The amplified luminescence proximity signal between AIM2-PYD-FLAG and AIM-HIN-200-Biotin was 940.0 ± 100.5 with no materials and 626.7 ± 98.7 upon incubation with poly(dA:dT) (Invivogen, San Diego, CA, USA). The difference of signals was significant (p = 0.000917) using Student’s t test (Fig. 2b). The data suggest that the PYD domain of AIM2 interacts with the HIN-200 domain of AIM2 loosely upon being incubated with no materials. Once the specific ligand poly(dA:dT) stringently interacts with the HIN-200 domain of AIM2, then the PYD domain of AIM2 apart from the HIN-200 domain of AIM2 is open to interact with the PYD domain of ASC. Since the PYD domain of ASC has been reported to loosely interact with the CARD domain of ASC [64], the interaction between the PYD domain of AIM2 and PYD domain of ASC is thought to open the CARD domain of ASC, which will lead to interaction with protein A-conjugated acceptor beads in the cell-free system (Fig. 3), or downstream CARD domain of caspase-1 in cells.
poly(dA:dT) reduces the amplified luminescent proximity signal between the PYD domain of AIM2 and the HIN-200 domain of AIM2. A schematic representation of full-length AIM2 (AIM2-FL) and its truncated proteins, the HIN200 domain of AIM2 (AIM2-HIN200) and PYD domain of AIM2 (AIM2-PYD). We synthesized two truncated forms of AIM2, AIM2-PYD-FLAG and AIM-HIN-200-Biotin, using a wheat germ cell-free synthesis system (a). Synthetic protein–protein interactions were detected by the amplified luminescent proximity homogeneous assay (Alpha). A total of 100 ng of each protein indicated was incubated with 5 μg/mL anti-FLAG mAb M2, 16.67 μg/mL protein-A-conjugated Alpha acceptor beads (PerkinElmer, Waltham, MA, USA), and 16.67 μg/mL streptavidin-conjugated Alpha donor beads (PerkinElmer, Waltham, MA, USA) for 24 h with or without 5 mg/mL poly(dA:dT) (Invivogen, San Diego, CA, USA). Responses (counts) were measured using EnSpire™ Multimode Plate Reader (PerkinElmer, Waltham, MA, USA). The results are given as means ± standard deviation from triplicate wells. Asterisk indicates significance (p < 0.01) (b)
A possible mechanism of AIM2 inflammasome in a cell-free system. The PYD domain of AIM2 interacts with the HIN-200 domain of AIM2 loosely upon being incubated with no materials. Once a specific ligand poly(dA:dT) stringently interacts with the HIN-200 domain of AIM2 and streptavidin-conjugated donor beads, then the PYD domain of AIM2 apart from the HIN-200 domain of AIM2 opens to interact with the PYD domain of ASC (a). Since the PYD domain of ASC loosely interacts with the CARD domain of ASC, the interaction between the PYD domain of AIM2 and PYD domain of ASC is thought to open the CARD domain of ASC, which will lead to interaction with protein A-conjugated acceptor beads (b)
Conclusions
Various inflammasomes are thought to play important roles in the maintenance of the homeostasis of cells, tissues, and organs. Excess inflammasome signaling caused by genetic mutations or pathogens may contribute to known or unknown autoinflammatory diseases. Thus, inflammasomes are expected to become attractive targets to treat autoinflammatory diseases. Although our cell-free system is limited in that only an initial event of assembly between ASC or RIPK2 and an upstream protein is detected, reconstituted inflammasomes in a cell-free system will be useful tools for investigating the pathogenesis of autoinflammatory diseases and discovery of their therapeutics.
References
Martinon F, Mayor A, Tschopp J. The inflammasomes: guardians of the body. Annu Rev Immunol. 2009;27:229–65.
Alnemri ES. Sensing cytoplasmic danger signals by the inflammasome. J Clin Immunol. 2010;30(4):512–9.
Franchi L, Eigenbrod T, Muñoz-Planillo R, Nuñez G. The inflammasome: a caspase-1-activation platform that regulates immune responses and disease pathogenesis. Nat Immunol. 2009;10(3):241–7.
Masters SL, Simon A, Aksentijevich I, Kastner DL. Horror autoinflammaticus: the molecular pathophysiology of autoinflammatory disease (*). Annu Rev Immunol. 2009;27:621–68.
Chen GY, Núñez G. Inflammasomes in intestinal inflammation and cancer. Gastroenterology. 2011;141(6):1986–99.
McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, Frucht DM, Aringer M, Torosyan Y, Teppo AM, Wilson M, Karaarslan HM, Wan Y, Todd I, Wood G, Schlimgen R, Kumarajeewa TR, Cooper SM, Vella JP, Amos CI, Mulley J, Quane KA, Molloy MG, Ranki A, Powell RJ, Hitman GA, O’Shea JJ, Kastner DL. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97(1):133–44.
Kastner DL, Aksentijevich I, Goldbach-Mansky R. Autoinflammatory disease reloaded: a clinical perspective. Cell. 2010;140(6):784–90.
Ombrello MJ, Kastner DL. Autoinflammation in 2010: expanding clinical spectrum and broadening therapeutic horizons. Nat Rev Rheumatol. 2011;7(2):82–4.
Pathak S, McDermott MF, Savic S. Autoinflammatory diseases: update on classification diagnosis and management. J Clin Pathol. 2017;70(1):1–8.
Srinivasula SM, Poyet JL, Razmara M, Datta P, Zhang Z, Alnemri ES. The PYRIN-CARD protein ASC is an activating adaptor for caspase-1. J Biol Chem. 2002;277(24):21119–22.
Martinon F, Burns K, Tschopp J. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-β. Mol Cell. 2002;10(2):417–26.
Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20(3):319–25.
Roberts TL, Idris A, Dunn JA, Kelly GM, Burnton CM, Hodgson S, Hardy LL, Garceau V, Sweet MJ, Ross IL, Hume DA, Stacey KJ. HIN-200 proteins regulate caspase activation in response to foreign cytoplasmic DNA. Science. 2009;323(5917):1057–60.
Hornung V, Ablasser A, Charrel-Dennis M, Bauernfeind F, Horvath G, Caffrey DR, Latz E, Fitzgerald KA. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458(7237):514–8.
Fernandes-Alnemri T, Yu JW, Datta P, Wu J, Alnemri ES. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature. 2009;458(7237):509–13.
Bürckstümmer T, Baumann C, Blüml S, Dixit E, Dürnberger G, Jahn H, Planyavsky M, Bilban M, Colinge J, Bennett KL, Superti-Furga G. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat Immunol. 2009;10(3):266–72.
Mariathasan S, Newton K, Monack DM, Vucic D, French DM, Lee WP, Roose-Girma M, Erickson S, Dixit VM. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature. 2004;430(6996):213–8.
Yu JW, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S, Sagara J, Fernandes-Alnemri T, Alnemri ES. Cryopyrin and pyrin activate caspase-1, but not NF-κB, via ASC oligomerization. Cell Death Differ. 2006;13(2):236–49.
Walsh JG, Muruve DA, Power C. Inflammasomes in the CNS. Nat Rev Neurosci. 2014;15(2):84–97.
Latz E, Xiao TS, Stutz A. Activation and regulation of the inflammasomes. Nat Rev Immunol. 2013;13(6):397–411.
Davis BK, Wen H, Ting JP. The inflammasome NLRs in immunity, inflammation, and associated diseases. Annu Rev Immunol. 2011;29:707–35.
Hoffman HM, Mueller JL, Broide DH, Wanderer AA, Kolodner RD. Mutation of a new gene encoding a putative pyrin-like protein causes familial cold autoinflammatory syndrome and Muckle-Wells syndrome. Nat Genet. 2001;29(3):301–5.
Aksentijevich I, Nowak M, Mallah M, Chae JJ, Watford WT, Hofmann SR, Stein L, Russo R, Goldsmith D, Dent P, Rosenberg HF, Austin F, Remmers EF, Balow Jr JE, Rosenzweig S, Komarow H, Shoham NG, Wood G, Jones J, Mangra N, Carrero H, Adams BS, Moore TL, Schikler K, Hoffman H, Lovell DJ, Lipnick R, Barron K, O’Shea JJ, Kastner DL, Goldbach-Mansky R. De novo CIAS1 mutations, cytokine activation, and evidence for genetic heterogeneity in patients with neonatal-onset multisystem inflammatory disease (NOMID): a new member of the expanding family of pyrin-associated autoinflammatory diseases. Arthritis Rheum. 2002;46(12):3340–8.
Feldmann J, Prieur AM, Quartier P, Berquin P, Certain S, Cortis E, Teillac-Hamel D, Fischer A, de Saint Basile G. Chronic infantile neurological cutaneous and articular syndrome is caused by mutations in CIAS1, a gene highly expressed in polymorphonuclear cells and chondrocytes. Am J Hum Genet. 2002;71(1):198–203.
Goldbach-Mansky R, Dailey NJ, Canna SW, Gelabert A, Jones J, Rubin BI, Kim HJ, Brewer C, Zalewski C, Wiggs E, Hill S, Turner ML, Karp BI, Aksentijevich I, Pucino F, Penzak SR, Haverkamp MH, Stein L, Adams BS, Moore TL, Fuhlbrigge RC, Shaham B, Jarvis JN, O’Neil K, Vehe RK, Beitz LO, Gardner G, Hannan WP, Warren RW, Horn W, Cole JL, Paul SM, Hawkins PN, Pham TH, Snyder C, Wesley RA, Hoffmann SC, Holland SM, Butman JA, Kastner DL. Neonatal-onset multisystem inflammatory disease responsive to interleukin-1β inhibition. N Engl J Med. 2006;355(6):581–92.
Gattorno M, Tassi S, Carta S, Delfino L, Ferlito F, Pelagatti MA, D’Osualdo A, Buoncompagni A, Alpigiani MG, Alessio M, Martini A, Rubartelli A. Pattern of interleukin-1β secretion in response to lipopolysaccharide and ATP before and after interleukin-1 blockade in patients with CIAS1 mutations. Arthritis Rheum. 2007;56(9):3138–48.
Hoffman HM, Brydges SD. Genetic and molecular basis of inflammasome-mediated disease. J Biol Chem. 2011;286(13):10889–96.
DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS, Trent JM. Cloning a novel member of the human interferon-inducible gene family associated with control of tumorigenicity in a model of human melanoma. Oncogene. 1997;15(4):453–7.
Man SM, Karki R, Kanneganti TD. AIM2 inflammasome in infection, cancer, and autoimmunity: role in DNA sensing, inflammation, and innate immunity. Eur J Immunol. 2016;46(2):269–80.
Miao EA, Alpuche-Aranda CM, Dors M, Clark AE, Bader MW, Miller SI, Aderem A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat Immunol. 2006;7(6):569–75.
Franchi L, Amer A, Body-Malapel M, Kanneganti TD, Ozören N, Jagirdar R, Inohara N, Vandenabeele P, Bertin J, Coyle A, Grant EP, Núñez G. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7(6):576–82.
Romberg N, Al Moussawi K, Nelson-Williams C, Stiegler AL, Loring E, Choi M, Overton J, Meffre E, Khokha MK, Huttner AJ, West B, Podoltsev NA, Boggon TJ, Kazmierczak BI, Lifton RP. Mutation of NLRC4 causes a syndrome of enterocolitis and autoinflammation. Nat Genet. 2014;46(10):1135–9.
Canna SW, de Jesus AA, Gouni S, Brooks SR, Marrero B, Liu Y, DiMattia MA, Zaal KJ, Sanchez GA, Kim H, Chapelle D, Plass N, Huang Y, Villarino AV, Biancotto A, Fleisher TA, Duncan JA, O’Shea JJ, Benseler S, Grom A, Deng Z, Laxer RM, Goldbach-Mansky R. An activating NLRC4 inflammasome mutation causes autoinflammation with recurrent macrophage activation syndrome. Nat Genet. 2014;46(10):1140–6.
Kitamura A, Sasaki Y, Abe T, Kano H, Yasutomo K. An inherited mutation in NLRC4 causes autoinflammation in human and mice. J Exp Med. 2014;211(12):2385–96.
The International FMF Consortium. Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial Mediterranean fever. The International FMF Consortium Cell. 1997;90(4):797–807.
Dowds TA, Masumoto J, Chen FF, Ogura Y, Inohara N, Núñez G. Regulation of cryopyrin/Pypaf1 signaling by pyrin, the familial Mediterranean fever gene product. Biochem Biophys Res Commun. 2003;302(3):575–80.
Masumoto J, Dowds TA, Schaner P, Chen FF, Ogura Y, Li M, Zhu L, Katsuyama T, Sagara J, Taniguchi S, Gumucio DL, Núñez G, Inohara N. ASC is an activating adaptor for NF-kappa B and caspase-8-dependent apoptosis. Biochem Biophys Res Commun. 2003;303(1):69–73.
Papin S, Cuenin S, Agostini L, Martinon F, Werner S, Beer HD, Grütter C, Grütter M, Tschopp J. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1β processing. Cell Death Differ. 2007;14(8):1457–66.
Mansfield E, Chae JJ, Komarow HD, Brotz TM, Frucht DM, Aksentijevich I, Kastner DL. The familial Mediterranean fever protein, pyrin, associates with microtubules and colocalizes with actin filaments. Blood. 2001;98(3):851–9.
Kim ML, Chae JJ, Park YH, De Nardo D, Stirzaker RA, Ko HJ, Tye H, Cengia L, DiRago L, Metcalf D, Roberts AW, Kastner DL, Lew AM, Lyras D, Kile BT, Croker BA, Masters SL. Aberrant actin depolymerization triggers the pyrin inflammasome and autoinflammatory disease that is dependent on IL-18, not IL-1β. J Exp Med. 2015;212(6):927–38.
Fukushima Y, Obara K, Hirata H, Sugiyama K, Fukuda T, Takabe K. Three Japanese patients (mother and two children) with familial Mediterranean fever associated with compound heterozygosity for L110P/E148Q/M694I and an autosomal true dominant inheritance pattern. Asian Pac J Allergy Immunol. 2013;31(4):325–9.
Stoffels M, Szperl A, Simon A, Netea MG, Plantinga TS, van Deuren M, Kamphuis S, Lachmann HJ, Cuppen E, Kloosterman WP, Frenkel J, van Diemen CC, Wijmenga C, van Gijn M, van der Meer JW. MEFV mutations affecting pyrin amino acid 577 cause autosomal dominant autoinflammatory disease. Ann Rheum Dis. 2014;73(2):455–61.
Marzano A, Damiani G, Ceccherini I, Berti E, Gattorno M, Cugno M. Autoinflammation in pyoderma gangrenosum and its syndromic form PASH. Br J Dermatol. 2016. doi:10.1111/bjd.15226 [Epub ahead of print].
Chamaillard M, Hashimoto M, Horie Y, Masumoto J, Qiu S, Saab L, Ogura Y, Kawasaki A, Fukase K, Kusumoto S, Valvano MA, Foster SJ, Mak TW, Nuñez G, Inohara N. An essential role for NOD1 in host recognition of bacterial peptidoglycan containing diaminopimelic acid. Nat Immunol. 2003;4(7):702–7.
Girardin SE, Boneca IG, Carneiro LA, Antignac A, Jéhanno M, Viala J, Tedin K, Taha MK, Labigne A, Zähringer U, Coyle AJ, DiStefano PS, Bertin J, Sansonetti PJ, Philpott DJ. Nod1 detects a unique muropeptide from gram-negative bacterial peptidoglycan. Science. 2003;300(5625):1584–7.
Inohara N, Ogura Y, Fontalba A, Gutierrez O, Pons F, Crespo J, Fukase K, Inamura S, Kusumoto S, Hashimoto M, Foster SJ, Moran AP, Fernandez-Luna JL, Nuñez G. Host recognition of bacterial muramyl dipeptide mediated through NOD2. Implications for Crohn’s disease. J Biol Chem. 2003;278(8):5509–12.
Girardin SE, Boneca IG, Viala J, Chamaillard M, Labigne A, Thomas G, Philpott DJ, Sansonetti PJ. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278(11):8869–72.
Kobayashi K, Inohara N, Hernandez LD, Galán JE, Núñez G, Janeway CA, Medzhitov R, Flavell RA. RIPK2/Rip2/CARDIAK mediates signalling for receptors of the innate and adaptive immune systems. Nature. 2002;416(6877):194–9.
Miceli-Richard C, Lesage S, Rybojad M, Prieur AM, Manouvrier-Hanu S, Häfner R, Chamaillard M, Zouali H, Thomas G, Hugot JP. CARD15 mutations in Blau syndrome. Nat Genet. 2001;29(1):19–20.
McGovern DP, Hysi P, Ahmad T, van Heel DA, Moffatt MF, Carey A, Cookson WO, Jewell DP. Association between a complex insertion/deletion polymorphism in NOD1 (CARD4) and susceptibility to inflammatory bowel disease. Hum Mol Genet. 2005;14(10):1245–50.
Weidinger S, Klopp N, Rummler L, Wagenpfeil S, Novak N, Baurecht HJ, Groer W, Darsow U, Heinrich J, Gauger A, Schafer T, Jakob T, Behrendt H, Wichmann HE, Ring J, Illig T. Association of NOD1 polymorphisms with atopic eczema and related phenotypes. J Allergy Clin Immunol. 2005;116(1):177–84.
Hysi P, Kabesch M, Moffatt MF, Schedel M, Carr D, Zhang Y, Boardman B, von Mutius E, Weiland SK, Leupold W, Fritzsch C, Klopp N, Musk AW, James A, Nunez G, Inohara N, Cookson WO. NOD1 variation, immunoglobulin E and asthma. Hum Mol Genet. 2005;14(7):935–41.
Plantinga TS, Fransen J, Knevel R, Netea MG, Zwerina J, Helsen MM, van der Meer JW, van Riel PL, Schett G, van der Helm-van Mil AH, van den Berg WB, Joosten LA. Role of NOD1 polymorphism in susceptibility and clinical progression of rheumatoid arthritis. Rheumatology (Oxford). 2013;52(5):806–14.
Madin K, Sawasaki T, Ogasawara T, Endo Y. A highly efficient and robust cell-free protein synthesis system prepared from wheat embryos: plants apparently contain a suicide system directed at ribosomes. Proc Natl Acad Sci U S A. 2000;97(2):559–64.
Masumoto J, Taniguchi S, Ayukawa K, Sarvotham H, Kishino T, Niikawa N, Hidaka E, Katsuyama T, Higuchi T, Sagara J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J Biol Chem. 1999;274(48):33835–8.
Kaneko N, Ito Y, Iwasaki T, Takeda H, Sawasaki T, Migita K, Agematsu K, Kawakami A, Morikawa S, Mokuda S, Kurata M, Masumoto J. Reconstituted AIM2 inflammasome in cell-free system. J Immunol Methods. 2015;426:76–81.
Dombrowski Y, Peric M, Koglin S, Kammerbauer C, Göss C, Anz D, Simanski M, Gläser R, Harder J, Hornung V, Gallo RL, Ruzicka T, Besch R, Schauber J. Cytosolic DNA triggers inflammasome activation in keratinocytes in psoriatic lesions. Sci Transl Med. 2011;3(82):82ra38.
Javierre BM, Fernandez AF, Richter J, Al-Shahrour F, Martin-Subero JI, Rodriguez-Ubreva J, Berdasco M, Fraga MF, O’Hanlon TP, Rider LG, Jacinto FV, Lopez-Longo FJ, Dopazo J, Forn M, Peinado MA, Carreño L, Sawalha AH, Harley JB, Siebert R, Esteller M, Miller FW, Ballestar E. Changes in the pattern of DNA methylation associate with twin discordance in systemic lupus erythematosus. Genome Res. 2010;20(2):170–9.
Dihlmann S, Erhart P, Mehrabi A, Nickkholgh A, Lasitschka F, Böckler D, Hakimi M. Increased expression and activation of absent in melanoma 2 inflammasome components in lymphocytic infiltrates of abdominal aortic aneurysms. Mol Med. 2014;20:230–7.
Zhen J, Zhang L, Pan J, Ma S, Yu X, Li X, Chen S, Du W. AIM2 mediates inflammation-associated renal damage in hepatitis B virus-associated glomerulonephritis by regulating caspase-1, IL-1β, and IL-18. Mediators Inflamm. 2014;2014:190860.
Dihlmann S, Tao S, Echterdiek F, Herpel E, Jansen L, Chang-Claude J, Brenner H, Hoffmeister M, Kloor M. Lack of absent in melanoma 2 (AIM2) expression in tumor cells is closely associated with poor survival in colorectal cancer patients. Int J Cancer. 2014;135(10):2387–96.
Ponomareva L, Liu H, Duan X, Dickerson E, Shen H, Panchanathan R, Choubey D. AIM2, an IFN-inducible cytosolic DNA sensor, in the development of benign prostate hyperplasia and prostate cancer. Mol Cancer Res. 2013;11(10):1193–202.
Iwasaki T, Kaneko N, Ito Y, Takeda H, Sawasaki T, Heike T, Migita K, Agematsu K, Kawakami A, Morikawa S, Mokuda S, Kurata M, Masumoto J. Nod2-nodosome in a cell-free system: implications in pathogenesis and drug discovery for Blau syndrome and early-onset sarcoidosis. ScientificWorldJournal. 2016;2016:2597376.
Masumoto J, Taniguchi S, Sagara J. Pyrin N-terminal homology domain- and caspase recruitment domain-dependent oligomerization of ASC. Biochem Biophys Res Commun. 2001;280(3):652–5.
Acknowledgements
Not applicable.
Funding
This work was supported by the Platform for Drug Discovery, Informatics and Structural Life Science from the Ministry of Education, Culture, Sports, Science and Technology, Japan (H.T., T.S., and J.M.), the Center for Clinical and Translational Research of Kyushu University (J.M.), and a Grant-in-Aid for translational research toward the clarification of autoinflammatory mechanisms by familial Mediterranean fever (FMF). Inflammasome based on the Mediterranean fever (MEFV) gene analysis 26310301 from The Ministry of Health, Labour and Welfare, Japan (J.M.), and Grants-in-Aid for Scientific Research (JSPS KAKENHI grant numbers 26293232 26305024 and 17H04656) from The Ministry of Education, Culture, Sports, Science and Technology, Japan (J.M.).
Availability of data and materials
Not applicable.
Authors’ contributions
The manuscript was written by all authors. Briefly, NK described about AIM2, TI described about Nod2, YI described about NLRP3. HT and TS synthesized proteins and described about wheat germ cell-free protein synthesis. SM, NN, MK drew figures. NK and JM described about inflammasome-related diseases and organized them. All authors read and approved the final manuscript.
Competing interests
All authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
Not applicable.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Kaneko, N., Iwasaki, T., Ito, Y. et al. Applications of reconstituted inflammasomes in a cell-free system to drug discovery and elucidation of the pathogenesis of autoinflammatory diseases. Inflamm Regener 37, 9 (2017). https://doi.org/10.1186/s41232-017-0040-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41232-017-0040-y