Abstract
Background
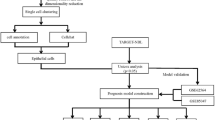
The Notch pathway combined with other signalling molecules acts specifically for the development of each blood cell type and differentiation stage. A causative role of Notch dysfunction in leukaemia development has been found in many studies so, initially only for T- acute lymphoblastic leukaemia (T-ALL) but more recently also for B cell and myeloid leukaemia. The aim of our study is to introduce a method for multiple direct analysis of the Notch pathway partners in a population of only 500 or fewer cells. The notion of this method consists in gaining insight into gene expression at the level of the malignant clone population. A small number of cells is a significant limitation when working on primary cells either when freshly isolated or when analysed after several days in cocultures.
Methods
The primers were designed to avoid genomic amplification through the selection of 3′ and 5′ primers that hybridise with different exons. Cell lines and primary cells were collected and multiplex quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR) performed on a descending number of cells, ranging from 2,500 cells up to 50 cells per sample, for the Notch pathway genes and other transcription factors important for cell differentiation. ImageJ program, STATISTICA 13.1 software package and Student’s t-test were used for statistical evaluation. We checked protein expression by western blot.
Results
We characterised the gene expression levels of Notch, Ikaros and Parp genes in leukaemia cell lines of B and T origin and in primary leukaemia samples of limited size. We further compared our results to the cDNA analysis obtained by total RNA isolation from a large number of cells as routinely performed in clinical laboratories, and finally tested the method described on primary cells from leukaemia patients.
Conclusions
This rapid multiple gene expression analysis of a small population of cells provides efficient cell classification determining malignant changes as an important additional information for clinical leukaemia diagnostics as well as for in vitro studies of primary cells.
Similar content being viewed by others
Background
The Notch family of genes is involved in the developmental routes and signalling pathways of many tissues across species and has very little diversity from insects to mammals [1]. However, its role is unique to the cell type, even if the cells are closely related, such as blood cells developing from the same hematopoietic stem cell. The variability of the Notch targets across different cells or cell differentiation step is also due to the ability to cooperate with other signalling pathways and to activate promoter sequences dependent on the chromatin status and actual cellular program. Recent investigations associate Notch signalling with super-enhancers, which serve as regulatory switches and lead to whole chromatin remodelling [2, 3]. Notch molecules and their ligands are cell transmembrane proteins. The signalling cascade begins with ligands binding to the Notch receptor in the cell membrane. In mammals there are three Delta-like ligands (Dll1, Dll3 and Dll4) and two Jagged ligands (JAG1 and JAG2), which can activate Notch receptors (NOTCH1, NOTCH2, NOTCH3 and NOTCH4). Notch receptors are first modulated through two cleavages by specific proteases, and then the ligand binding causes a third proteolytic cleavage via γ-secretase, the subsequent release of the Notch intracellular domain (NICD) and its nuclear relocation. In the nucleus this molecule forms a large complex interacting with cofactors, such as the transcription factor complex CSL (CBF1/Su (H)/Lag1), which binds to a specific sequence, displaces a repressor complex and activates transcription. In addition, a number of interacting molecules, from modulators of ligand binding to Notch inhibitors and competing factors, influence the outcome of this signalling [1, 4]. Feedback loops regulate the signalling length and may also have a role in establishing the specificity of the response.
In haematopoiesis, Notch signalling is one of the major regulators not only of lymphocyte but also myeloid development. It regulates several controlling steps in hematopoietic cell differentiation and is responsible for T cell commitment from the multipotent precursor while blocking early B cell lymphopoesis. It is involved in several checkpoints in thymic T cell development and peripheral T cell differentiation [5,6,7], in the specification of B cell subsets in the spleen and also for dendritic cell function [8].
Notch was first proposed to have a causative role in leukaemia development in 1996 [9]. Notch mutations or chromosome translocations leading to hyper activation are evident in nearly 60% of T cell acute lymphoblastic leukaemia (T-ALL) patients [10, 11]. While Notch activation in B cell chronic lymphocytic leukaemia could lead to apoptosis and growth arrest, in B cell chronic lymphocytic leukaemia and several types of lymphomas, its mutation and subsequent pathway activation can lead to more aggressive forms of the disease [12,13,14]. In multiple myeloma, the appearance of plasma cell malignancy and Notch activation strongly depend on the receptor interaction with its ligands in the microenvironment [11]. The accumulation of data on the importance of Notch in the process of leukemogenesis also led to the first clinical trials where Notch inhibitors were used in the treatment of T-ALL [15].
In this article, we characterised leukaemia cell lines of B and T origin as well as primary leukaemia cells, according to the expression of upstream and downstream elements of the Notch signalling pathway. This type of data could, depending on the cell type, contribute to the development of more precise prognostic markers and tailored therapies. We analysed gene expression by qRT-PCR using either cDNA obtained from purified RNA which usually requires at least one million cells or by performing reverse transcription directly without previous RNA purification. We titrated down the necessary cells for the former to a very small number (five to 500). Therefore this method of rapid gene expression analysis is applicable for an efficient classification of cells and the determination of malignant changes that could serve as potential diagnostic markers in samples as small as 10 μl of peripheral blood.
Methods
Primer design
All data on eligible gene sequences were obtained from the Ensembl Project database. All primers were designed using publicly available software, such as Primer3Plus and IDT Primer Quest Tool. For each gene of interest three primers were constructed, two forward (F1 and F2) and one reverse (R). The primers were designed to avoid genomic amplification through the selection of 3′ and 5′ primers that hybridise with different exons. To ensure that the primers had the same amplification efficiency, we attempted to design them to have a similar number of base pairs and similar melting temperatures (Tm), a GC content around 50% and a maximum ΔG value of the five bases from the 3′ end. The length of the expected amplicon of the first polymerase chain reaction (PCR) did not exceed 330 base pairs (bp), while the length of the amplicon of the second PCR did not exceed 200 bp. The uniqueness of binding to the human genome was checked in BLAST. All primers were verified to not create homodimers, and it was ensured that those used in the first PCR did not create heterodimers. Primer compatibility and the size of the amplified fragments were checked using the freely available software program AmplifyX. PCR with primer pairs in the first and second PCR were compared to determine the equal primer efficacy.
Patients, cell lines and cell cultures
Cell lines were obtained from the German Collection of Microorganisms and Cell Cultures, including Jurkat (human T cell leukaemia cells), CLL (B chronic lymphocytic leukaemia cells); and 697 and Nalm 6 (both human B cell precursor leukaemia cells). Cells were cultured in RPMI-1640 medium supplemented with 10% foetal calf serum (FCS, Sigma–Aldrich). Primary cells were collected from peripheral blood of two newly diagnosed patients. Patients were diagnosed with chronic lymphoblastic leukaemia according to the World Health Organisation (WHO) classification of haematolymphoid malignancies using combined diagnostic information from cytomorphology, cytochemistry, immunophenotyping, immunogenetics and molecular cytogenetics characteristics. Informed consent and Ethics Review Board approval by the Ethics Committee of the University of Zagreb Medical School was obtained, and the experiments were performed according to the Declaration of Helsinki.
RNA isolation and preparation of cDNA
Total RNA was extracted using TRI Reagent (Sigma–Aldrich) according to the manufacturer’s instructions. The integrity and purity of isolated RNA was checked on agarose gel by electrophoresis, measured on NanoVue (GE Life Sciences) and 2 μg of RNA were retro-transcribed using random hexamers (Roche) and PrimeScript (TaKaRa).
Direct reverse transcription from cell lysates and PCR amplification
Cells were harvested when optimal density was reached, rinsed twice with phosphate buffer saline, counted and finally aliquoted. The multiplex PCR method was performed as described by Peixioto [16], but with modifications we introduced in order to use 50 to 2,500 cells, instead of single cells as originally described. The cells were first snap frozen at − 80 °C, then heated to 65 °C for two minutes, followed by 50 min at 42 °C, with MuLV Reverse Transcriptase and 100 nM of 3′ gene specific primers from Applied Biosystems. The product obtained was used as a template in the first PCR amplification, with 15 nM of F1 and R primer pair and with Taq polymerase (Sigma–Aldrich). In the first and second semi nested PCR with F2 and R primer pair, cDNA was amplified for 15 and 35 cycles respectively. We designed the primers for expression analysis by semi-nested PCR for the amplification of NOTCH1, NOTCH2, NOTCH3, Dll1, JAG1, Hes-1, Hey-1, NUMB, Aiolos (IKZF3), PARP-1, PARG, RUNX, PAX-5 and PU.1 and for HPRT (hypoxanthine phosphoribosyl transferase) as a housekeeping gene. In addition to that, a literature search confirmed that primers for NOTCH2 and JAG1 were previously successfully applied [17, 18].
qRT-PCR
qRT-PCR was performed using a 7500 Fast Real-Time PCR System in a 96-well plate. Each well contained 1X SYBR Green PCR MASTER MIX (all reagents and instruments from Applied Biosystems), 150 nM of primers and 1 μl of template, either from the first PCR amplification where cDNA was obtained from total cells or from cDNA obtained from purified RNA. PCR efficiency was calculated from the slope of the amplification curve in the exponential phase by means of linear regression software and was higher than 90%. Melting curve analysis was always used for determination of product specificity.
Western blotting
Total cell extracts were prepared using a lysis buffer containing a cocktail of protease inhibitors, as previously described [18]. Protein concentration was measured according to Bradford and analysed using the western blot chemiluminescence detection method. The primary antibodies used were rabbit anti human-Notch2, rabbit anti human-Hes1 (Cell Signalling Technology, USA), rabbit anti human-PARP-1 and rabbit anti human-β-actin (Santa Cruz).
Statistical analysis
The ImageJ program (provided by the National Institute of Health, Bethesda, USA) was applied for densitometric analysis. Statistical analysis was performed using the STATISTICA 13.1 software package (Stat Soft Inc., USA). Student’s t-test was used for comparison of results gained by the two different methods. The significance level was set at p < 0.05.
Results
Transcription factor expression in human leukaemic cells of B and T origin by RT-PCR
In the developmental decisions of B and T lymphogenesis, as well as in the development of certain types of leukaemia, Notch signalling plays an important role. Therefore, we analysed the different components of the Notch pathway in four human leukaemia-derived cell lines. We selected representative samples of a T cell leukaemia (Jurkat), pre-B cell leukaemia (697 and Nalm 6), and B chronic lymphocytic leukaemia (CLL) and analysed the expression of 14 genes via qRT-PCR. Most of the genes we studied are involved in Notch signalling as receptors (NOTCH-1, − 2, − 3), ligands (Dll1, JAG1) or downstream effectors (HES-1, HEY-1) as well as inhibitors (NUMB). Further, we analysed Aiolos (IKZF3), PARP-1, PARG, RUNX, PAX-5 and PU.1, all of which are involved in lymphocyte differentiation and development (see Fig. 1).
Validation of primer design strategy. a Efficiency: PCR reactions amplifying cDNA fragments with F1 and F2 semi-nested primers should have the same efficiency. The efficiency of the designed primers was tested on the mixture of all cell line cDNA samples. b Competition: 12 primers present in the same reaction should not interfere with each other. After the first multiplex PCR reaction using the cDNA mixture as a template, separate PCR reactions were set on with semi-nested F2 primers to analyse the specificity of the reaction. M (100 bp DNA Ladder), all amplicons cited with the name of the gene; longer amplicons produced by F1/R and shorter by F2/R primers
Relative expression of Notch and PARP pathways are presented on Fig. 2. We found that pre-B cells, 697 and Nalm-6, expressed NOTCH2 but very low levels of NOTCH1 and NOTCH3. Genes dependent on Notch signalling pathway activation, such as HES1 and HEY, were not expressed, and transcripts of only one ligand type, JAG1, were present. On the other hand, genes that determine B cell development, PAX-5 and PU.1, were active. The expression of Aiolos, RUNX, PARP-1, PARG and NUMB was high in comparison with that of the reference gene. The cell line, representing B chronic lymphocytic leukaemia cells (CLL) (see Fig. 2), showed a high expression of NOTCH1 and NOTCH 2 but not NOTCH3. These cells also expressed the downstream effectors of Notch signalling, HES1 and HEY1. JAG1 ligand expression was also detected. Originating from B cells, these cells also had PAX-5 and PU.1 transcription factor expression (Fig. 2). The T cell leukaemia cells (Jurkat) expressed NOTCH1, NOTCH 2, and NOTCH 3 and JAG1 receptors as well as downstream effectors of active Notch signalling, HES1 and HEY1, with only a modest expression of the Dll1 ligand. The expression of Aiolos was lower than what was found in B cells. RUNX was highly expressed in all leukaemia samples. B cell gene characteristics were not expressed, such as the B lymphocyte-developmental master transcription factor PAX-5 and PU.1. Transcription of the genes involved in PARP-1 regulation, PARP-1 and PARG, was also detected. For further analysis we have chosen NOTCH2, PARP-1, HES-1, PAX-5 and Aiolos because of their differential expression in the leukemic cells tested in Fig.2. and their possible applicability as a fast diagnostic tool.
Relative expression of Notch (upper panel) and PARP (lower panel) pathways by RT-PCR in cell lines of B and T cell origin (cDNA obtained from Jurkat, 697, Nalm-6 and CLL cell lines). Relative expression by band densitometry after gel electrophoresis of PCR amplicons, normalised with the level of HPRT expression. M ± SD of two independent experiments
Relative quantification of transcription factors via multiplex qRT-PCR
In order to achieve transcription factor expression at the cell population level, challenging the more sophisticated and expensive single cell multiplex qRT-PCR and seeking to reduce the number of cells necessary when samples are derived from patients, we established a new fast, simple and reliable method for gene expression analysis. We conceived a modification of the single cell multiplex RT-PCR method of Peixoto et al. [16]. In the first step we constructed the primers according to a number of conditions, as previously described. For each gene of interest three primers were constructed, two forward (F1 and F2) and one reverse (R), to obtain better specificity in a semi-nested PCR reaction. From our RT-PCR results, we chose differentially expressed genes across cell lines of B and T lymphocyte origin and also genes that could tell us more about the cell differentiation status. Used together in the first multiplex PCR reaction, F1 and R primers from various genes did not mutually interact (as shown in a competition test in Fig. 1b). Further, to obtain reliable quantitative relations, the efficiency of the F1 and F2 primers was shown to be equal (Fig. 1). We prepared lysates from leukemic cells ranging from 50 to 2,500 cells and performed direct reverse transcription initiated by a set of gene-specific primers (R). The obtained cDNA was first amplified in a multiplex PCR reaction and then in separate, semi-nested reactions. The results obtained from samples ranging from 50 to 2,500 cells revealed no significant inhibition of the PCR reaction originating from higher cell number extracts (Fig. 3a). Since the samples with 50 cells had slightly more variability we selected the samples with 500 cells for further analysis. The 500-cell samples proved to be just right to avoid differences among single cell expression levels while simplifying the manipulation steps, and yet the number is small enough not to inhibit the PCR reaction and to be obtainable from less than 10 μl blood for several analyses.
a) Quantitative analysis of gene expression in samples obtained from cell lysates of different numbers of 697 cells. PCRI: cDNA obtained by direct reverse transcription from lysates of 2,500; 1,500; 500 and 50 cells. b) Quantitative analysis of gene expression in samples obtained from reverse transcription of total RNA isolate (■ cDNA) and from cell lysates (□ PCRI) of 697 cells. Relative expression was presented as ΔCt value normalised with HPRT gene expression. ΔCt value of 10 means that there was no amplification. The results presented were obtained from three different experiments and shown as mean ± SD. Differences between the means was not statistically significant (p > 0.05). cDNA: cDNA obtained from RNA isolated from 2 × 106 cells; PCR I: cDNA obtained by direct reverse transcription of lysate from 500 cells
Figure 3b represents the comparison of gene expression levels obtained by cDNA amplification. In the first set of samples, the template was cDNA produced by reverse transcription of isolated total RNA (cDNA). In the second set, the template was derived by direct reverse transcription of RNA in cell lysates and subsequent 15 cycles amplification of multiplex PCR reaction (PCRI). The gene expression level is presented as a Delta Ct (Cycle threshold) using the HPRT expression as a reference value. Although there were slight differences in the levels of expression between different templates, the relationship between the levels of expression of various genes was unchanged throughout all the experiments. The analysed cells were 697, pre-B lymphocyte strain cells, which showed a low expression of HES-1, a downstream effector of Notch signalling, as expected in pre-B cells. Notch-2 and Aiolos were expressed moderately, while PARP-1 and Pax-5 showed significant expression. Gene expression was also compared using the PARP-1 expression level in order to show that direct analysis preserved the same expression ratio as classical reverse transcription cDNA.
Next, we applied this method to characterise the signalling pathways in three additional lymphocyte cell lines. As the primary purpose for developing this method was to analyse a small number of cells in samples obtained from patients, we also demonstrated the applicability of the method on patient samples. Fig. 4 presents the analysis of Notch- and PARP-1-related pathways in different cells of B and T origin. Similar to 697 pre-B cells, Nalm6 showed a very low expression of Hes1, which indicates the absence of Notch activity. The cells also expressed low levels of Notch2 receptor. PARP-1 and Aiolos were highly expressed, as was Pax-5, a B cell transcription marker. Jurkat cells, on the contrary, demonstrated an increase in Hes expression, a feature indicating an active Notch pathway, which is characteristic of T lymphocyte development. These cells expressed a higher level of the Notch2 receptor when compared to 697 cells, but the B cell marker Pax-5, as expected, was not present at all. Cells obtained from the CLL line expressed Hes1, showing a higher activity of the Notch pathway. Notch2 receptor and Pax-5 were moderately expressed. The analysis of primary leukaemia cells showed that this method is also applicable to lymphocytes previously separated from blood samples.
Quantitative gene expression analysis in four human-derived cell lines and primary cells obtained from two patients (Sample 1 and 2). Cell lysates obtained from 500 cells were reversely transcribed with gene-specific primers and amplified for 15 cycles in multiple PCR reaction. For qRT-PCR reaction, the primer pairs were semi-nested. Expression levels are presented as ΔCt values
We also analysed the expression of Notch downstream effectors and PARP signalling proteins (Fig. 5). Western blot analysis of the cell lines tested confirmed the data obtained by qRT-PCR. In Jurkat cells, Notch2 receptor (FL) was expressed as well as its downstream target Hes1, indicating the activity of the Notch pathway. PARP-1 showed moderate expression. In the 697 cells, Notch2 N terminal form (NTM) was highly expressed, but no Hes-1 protein was detected. PARP-1 showed higher expression levels in 697 cells in comparison with other lines, similar to the results obtained by qRT-PCR. Nalm6 cells also showed Notch-2 expression, but no downstream activity leading to Hes1 expression was detected. In CLL cells, Notch2 receptor was expressed, but so was Hes1, indicating downstream Notch signalling activity. PARP-1 showed a lower level of expression in comparison to pre-B cells.
Discussion
Certain genes interfere with Notch signalling at promoter (such as Aiolos) [19, 20] and PARP-1 levels [21, 22]. PARG interferes with PARP-1 pathways and could be indirectly involved in Notch signalling. Runx is a transcription factor involved in blood cell development [23], as are Pax-5 and Pu.1, which are characteristic master transcription factors directing B cell development [23,24,25].
The genes analysed were those involved in Notch signalling (Hes1, Notch2), Pax-5 as a B cell marker, PARP-1 and Aiolos, an Ikaros family member involved in B cell differentiation. Notch2 was chosen as the most expressed Notch receptor in different cell lines and Hes-1 as an indicator of Notch pathway activity. Therefore, in this article, we analysed the expression of genes involved in lymphocyte differentiation in several B and T leukaemia cell lines as well as in samples obtained from leukaemia patients. The focus was on the Notch signalling pathway as one of the main pathways determining the diversification between B and T cells.
The 697 and Nalm-6 pre-B cell lines showed typical expression of B cell origin genes: the presence of Pax-5 and Pu.1 and the absence of an active Notch pathway, as demonstrated by the repression of downstream targets of cleaved NOTCH, HES and HEY genes. It was found that cells do express Notch receptors, with Notch-2 as the dominant form, as well as some ligands (such as JAG1). Inactive Notch signalling was also confirmed at the protein level. In general, B cells during development show low Notch signalling activity, although Notch was found to have a role in marginal zone B cells.
Another B cell line, CLL, showed, in addition to the expression of Notch receptors and low levels of ligands, the expression of all Notch downstream targets we analysed, indicating signalling activity. Hes-1 protein was also detected at the protein level. This result is in accordance with the recently described subset of CLL, where a mutation in the Notch-1 PEST domain increases Notch half-life and contributes to the aggressiveness of the disease [26, 27]. The detection of downstream Notch targets indicates the role of this pathway in the aetiology of CLL.
Jurkat cells have an active Notch pathway at the RNA and protein levels. These data are in concordance with the known role of Notch in T cell differentiation. Constitutive Notch signalling is involved in T cell malignancies, where it interferes with differentiation, proliferation and survival [28]. In order to obtain insight into the gene expression of the malignant clone at the population level we utilised the method described by Peixioto et al. [16] for single cells but in a population of cells as small as 500 for multiple direct analysis of the Notch pathway partners. A small number of cells is a significant limitation when working on primary cells, especially when dealing with very small samples of tissue or paediatric samples. The method provided the same results as PCR from isolated total RNA. This newly introduced method is a short-cut technique for detecting key Notch and Parp gene expression. Namely, the development of new technologies enables scientists to analyse RNA expression at the level of one cell, and even to detect the existence of only two molecules of DNA. These methods are valuable when only a small number, or only one cell, is available, such as in the case of early embryonic cells. Moreover, these methods make the analysis of differences among cells in a cell population possible. Such analyses from several studies indicate the existence of variability among gene expression profiles in cells of the same type within a population. At the same time, these techniques require laborious validation, as the resultant values originate from the unique cell, and only the reliability of the method proves the diversity among entities. This variability in gene expression among single cells is avoided when analysing several hundred cells at the same time [17]. Our method has been proven to be sensitive, and the results obtained are in concordance with those of methods involving the RNA isolation step as well as with protein analysis. Multiplex PCR reaction enables amplification of rare sequences, avoiding their loss. Small differences that appear in expression levels between samples obtained in different ways likely stem from two-step PCR reactions applied in direct analysis. Differences also occur when different types of reverse transcription reactions are applied, i.e. depending on the primers used (random or specific primers or oligo dT) (data not shown). It should be emphasised that primer design is the most critical step. Although all primer pairs were checked for their efficiency, it is still possible that small differences existed. On the other hand, it is conceivable that the presence of other components of cell lysates influenced the results. Nonetheless, the ratio of gene expression in the multiplex reaction was preserved when compared with cDNA amplification. The advantages of this method are, besides the small number of cells necessary for the tests, the simple, straight-forward sample preparation, direct approach and simultaneous analysis of the desired set of genes. In addition, along with cell sorting it enables the analysis of small populations of cells (e.g. stem cells) in small biological samples. These features are especially important in conditions when only small samples are available, such as in paediatric samples, as well as when fast and simple diagnostic methods are required. Also, our method is relevant for a laboratory where only few cell culture cells are available that cannot be processed through a cell sorter or for a medical centre where a fast reliable result is required in spite of a sophisticated cell sorter facility. Uncovering the underlying processes that contribute to the leukemic transformation or the maintenance of leukemic cells could potentially lead to the development of more efficient and precise therapeutic strategies.
Further the results presented here regarding Notch signalling analysis in leukaemia samples could contribute to efficient and person-oriented therapy in the future.
Conclusion
In this article, we characterise leukaemia cell lines of B and T origin, as well as primary leukaemia cells, according to the expression of upstream and downstream elements of the Notch signalling pathway. For this work, we introduced a newly designed method of rapid expression analysis applicable for the efficient classification of cells and determination of malignant changes that could serve as potential markers in leukaemia diagnostics.
Abbreviations
- B-CLL:
-
B - Chronic lymphocytic leukaemia cells
- bp:
-
Base pairs
- CBF1/Su (H)/Lag1:
-
Transcription factor complex CSL
- CLL:
-
Name of the cell line representing B chronic lymphocytic leukaemia cells
- Dll1:
-
Delta-like ligands
- FCS:
-
Foetal calf serum
- HPRT:
-
Hypoxanthine phosphoribosyl transferase
- JAG:
-
Jagged ligands
- NICD:
-
Notch intracellular domain
- NTM:
-
NOTCH2 N terminal form
- qRT-PCR:
-
Quantitative real-time reverse transcription polymerase chain reaction
- T-ALL:
-
T- Acute lymphoblastic leukaemia
References
Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signalling: cell fate control and signal integration in development. Science. 1999;284:770–6.
Somasundaram R, Prasad MAJ, Ungerbäck J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 2015;126(2):144–52.
Wang H, Zang C, Taing L, Arnett KL, Wong YJ, Pear WS, et al. NOTCH1-RBPJ complexes drive target gene expression through dynamic interactions with superenhancers. Proc Natl Acad Sci. 2014;111(2):705–10.
Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13:654.
Taghon TN, David E-S, Zuniga-Pflucker JC, Rothenberg EV. Delayed, asynchronous, and reversible T-lineage specification induced by Notch/Delta signaling. Genes Dev. 2005;19(8):965–78.
Osborne BA, Minter LM. Notch signalling during peripheral T-cell activation and differentiation. Nat Rev Immunol. 2007;7:64.
Koyanagi A, Sekine C, Yagita H. Expression of Notch receptors and ligands on immature and mature T cells. Biochem Biophys Res Commun. 2012;418(4):799–805.
Radtke F, MacDonald HR, Tacchini-Cottier F. Regulation of innate and adaptive immunity by Notch. Nat Rev Immunol. 2013;13:427.
Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183(5):2283–91.
Breit S, Stanulla M, Flohr T, Schrappe M, Ludwig W-D, Tolle G, et al. Activating NOTCH1 mutations predict favorable early treatment response and long-term outcome in childhood precursor T-cell lymphoblastic leukemia. Blood. 2006;108(4):1151–7.
Mirandola L, Comi P, Cobos E, Martin Kast W, Chiriva-Internati M, Chiaramonte R. Notch-ing from T-cell to B-cell lymphoid malignancies. Cancer Lett. 2011;308(1):1–13.
Tohda S, Kogoshi H, Murakami N, Sakano S, Nara N. Diverse effects of the Notch ligands Jagged1 and Delta1 on the growth and differentiation of primary acute myeloblastic leukemia cells. Exp Hematol. 2005;33(5):558–63.
Fabbri G, Rasi S, Rossi D, Trifonov V, Khiabanian H, Ma J, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389–401.
Hernandez Tejada FN, Galvez Silva JR, Zweidler-McKay PA. The challenge of targeting Notch in hematologic malignancies. Front Pediatr. 2014;2(54) https://doi.org/10.3389/fped.2014.00054.
Andersson ER, Lendahl U. Therapeutic modulation of Notch signalling — are we there yet? Nat Rev Drug Discov 2014; 13:357.
Peixoto A, Monteiro M, Rocha B, Veiga-Fernandes H. Quantification of multiple gene expression in individual cells. Genome Res. 2004;14:1938–47.
Sciacca S, Pilato M, Mazzoccoli G, Pazienza V, Vinciguerra M. Anti-correlation between longevity gene SirT1 and Notch signaling in ascending aorta biopsies from patients with bicuspid aortic valve disease. Heart Vessel. 2013;28(2):268–75.
Hayashi Y, Osanai M, Lee G-H. NOTCH2 signaling confers immature morphology and aggressiveness in human hepatocellular carcinoma cells. Oncol Rep. 2015;34(4):1650–8.
Koipally J, Renold A, Kim J, Georgopoulos K. Repression by Ikaros and Aiolos is mediated through histone deacetylase complexes. EMBO J. 1999;18(11):3090–100.
Kathrein KL, Chari S, Winandy S. Ikaros directly represses the Notch target gene Hes1 in a leukemia T cell line: implications for CD4 regulation. J Biol Chem. 2008;283(16):10476–84.
Gulino A, Di Marcotullio L, Screpanti I. The multiple functions of Numb. Exp Cell Res. 2010;316(6):900–6.
Kannan S, Fang W, Song G, Mullighan CG, Hammitt R, McMurray J, et al. Notch/HES1-mediated PARP1 activation: a cell type-specific mechanism for tumor suppression. Blood. 2009;117(10):2891–900.
McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, et al. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15(20):5647–58.
Ernst P, Fisher JK, Avery W, Wade S, Foy D, Korsmeyer SJ. Definitive hematopoiesis requires the mixed-lineage leukemia gene. Dev Cell. 2004;6(3):437–43.
Cs C, Schebesta A, Delogu A, Busslinger M. Pax5: the guardian of B cell identity and function. Nat Immunol. 2007;8:463.
Rosati E, Sabatini R, Rampino G, Tabilio A, Di Ianni M, Fettucciari K, et al. Constitutively activated Notch signaling is involved in survival and apoptosis resistance of B-CLL cells. Blood. 2009;113(4):856–65.
Gianfelici V. Activation of the NOTCH1 pathway in chronic lymphocytic leukemia. Haematologica. 2012;97(3):328–30.
Weng AP, Ferrando AA, Lee W, Morris JP, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306(5694):269–71.
Acknowledgements
The authors wish to thank Dr. Lipa Čičin-Šain for figure preparation, the associations and all the candidates who volunteered for this study.
Funding
This project was made possible by funding received from the Croatian Science Foundation grant IP-11-2013 7140 (Notch in hematopoietic stem cell differentiation and leukaemia development); The Terry Fox Foundation and Croatian League Against Cancer; the European Union’s Seventh Programme for research, technological development and demonstration under grant agreement No. 602587 (Development of Stem Cell Based Therapy for Thymic Regeneration); as well as the Ministry of Science and Education of the Republic of Croatia.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
MA and DR-K designed the study, analysed and interpreted the results and wrote the manuscript; LH, JS, IF, DH and JM carried out the experiments, analysed the samples and co-wrote the manuscript; MM, IK-S and BJ-P critically revised the data and their implementation in a future diagnostics and interpretation of results and co-wrote the paper. All the authors have accepted responsibility for the entire content of this submitted manuscript and approved submission.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Patients’ informed consent and Ethics Review Board approval by the Ethics Committee of the University of Zagreb Medical School was obtained, and the experiments were performed according to the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Horvat, L., Skelin, J., Puškarić, B.J. et al. Notch pathway connections in primary leukaemia samples of limited size. transl med commun 3, 8 (2018). https://doi.org/10.1186/s41231-018-0026-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s41231-018-0026-z