Abstract
Background
Type 2 diabetes mellitus is a multi-factorial disease in which both genetic and non-genetic factors interact in order to precipitate the diabetic phenotype. Among various predisposing genetic loci, a pentanucleotide (CTTTA) Del/Ins variant in the 3'-UTR of the LEPR gene is associated with type 2 diabetes and its related traits. This study was done to explicate for the first time the association of this Del/Ins polymorphism of LEPR gene in type 2 diabetes patients belonging to the ethnic population of Kashmir valley.
Methods
670 unrelated subjects comprising of 320 type 2 diabetes patients and 350 healthy controls were included in the study. Genotyping of the untranslated region of LEPR gene encompassing this Del/Ins variant was done by PCR-RFLP technique and results were validated by direct sequencing.
Results
Genotype frequencies for both type 2 diabetes cases and healthy controls were consistent with Hardy-Weinberg equilibrium (χ2 = 3.09 and 2.37, P = NS). The Del/Del genotype was predominantly found in cases than controls (P = 0.003, OR: 0.62, CI: 0.45-0.85). Carriers of Ins/Ins genotype were relatively protected against the risk factors (P = 0.0004, OR: 0.31, 95% CI: 0.15-0.61). A positive association was observed between the Del allele and the risk factors of type 2 diabetes.
Conclusion
The results elucidate that the CTTTA Del allele is a genotypic risk factor of type 2 diabetes in the Kashmiri population.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
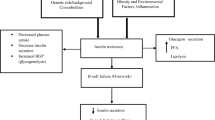
Type 2 Diabetes Mellitus (T2DM) is a multifarious disorder caused by interactions between susceptible genetic loci and environmental slurs. T2DM is an incipient non-communicable global epidemic [1]. The genetics of T2DM is incoherent with respect to the patterns of simple Mendelian inheritance. Contemporary studies inspecting the genetic elements involved in the development of T2DM have not identified any disease-causing mutations; however several predisposing genetic variants (polymorphisms) have been recognized [2]–[4]. The heterogeneity of this disorder renders the understanding of risk factors and sequence of events obscure. Currently the involvement of diverse proteins produced and secreted by adipocytes, collectively termed as adipokines is recognized to modulate insulin resistance [5]. Amongst these adipokines; leptin, resistin, TNF-α and free fatty acids are implicated in convening insulin resistance whereas adiponectin improves sensitivity. Inconsistency between insulin resistant and insulin sensitizing adipokines is implicated in type 2 diabetes and related disorders. Leptin exerts its central and peripheral effects by binding to its receptors that belong to the class I cytokine receptor superfamily [6]. This binding results in subsequent activation of downstream signalling pathways regulating satiety and stimulating energy expenditure [7]. Sequential phenotypic manifestations of diabetes like hyperinsulinemia, obesity and β-cell failure are observed in animal models (ob/ob and db/db mice) [8],[9]. Disparity between leptin-insulin alliance in pancreatic β-cells as a result of leptin resistance in overweight patients has been suggested to promote hyperinsulinemia and subsequent development into T2DM [10],[11]. In pancreatic β-cells, inhibition of insulin secretion by leptin has been shown to be mediated by the long form of LEPR gene [12],[13]. The human leptin receptor gene (LEPR) is localised in proximity of the microsatellite marker D1S198 which is associated with acute insulin response [14]–[16]. The 3'-UTR (untranslated region) of LEPR gene encompasses a pentanucleotide (CTTTA) Deletion/Insertion polymorphism. Studies investigating this Del/Ins polymorphism have shown an association with susceptibility to type 2 diabetes and related risk factors [15],[17]–[22]. We screened the CTTTA-pentanucleotide Del/Ins in 3'-UTR of LEPR gene in ethnic Kashmiri population of north India. Our study is the first ever molecular characterization of type 2 diabetes mellitus patients of Kashmiri valley aimed to study a possible association between this genetic polymorphism and type 2 diabetes mellitus.
Methods
Study population and Clinical parameters
The study included 670 ethnic Kashmiri subjects, comprising of 320 unrelated type 2 diabetes mellitus patients and 350 healthy controls assenting to participate voluntarily. Type 2 diabetes cases were diagnosed as per ADA/WHO criteria. Controls were matched for age and gender. Exclusion criteria involved patients with drug induced, stress induced and gestational diabetes. The study was approved by institutional ethical committee (IEC-SKIMS) and the research protocol conforms to the provisions of the Declaration of Helsinki (as revised in Tokyo 2004). 4 ml of blood was collected from the cubital vein after 12 hours of fast from each subject. Serum was separated from 2 ml of blood and immediately sent for biochemical analyses. Remaining 2 ml of blood was stored in EDTA vacutainer at −70°C until processed for isolation of DNA. Laboratory tests including fasting plasma glucose (FPG), total cholesterol (TC), triglycerides (TG), high density lipoprotein (HDL), low density lipoprotein (LDL) and serum creatinine levels were measured on an autoanalyzer.
Hypertension was defined as a systolic pressure ≥140 mmHg or diastolic pressure ≥90 mmHg. Blood pressure (BP) was measured using standard sphygmomanometer in the sitting position after a 10 minute rest. Anthropometric data comprised of Body Mass Index (BMI) and Waist to Hip ratio (WHR). Height and weight were measured using standard anthropometric techniques in light-weight clothing without shoes. Waist circumference was measured midway between the lower rib margin and the iliac crest at the end of gentle expiration using an anthropometric measuring tape. BMI was expressed as kg/m2.
Genotyping
DNA was extracted from peripheral leukocytes by Qiamp Blood Mini extraction kit (Qiagen Valencia, CA). The variant CTTTA pentanucleotide insertion was screened by polymerase chain reaction (PCR) by amplifying a 114/119 bp region of LEPR gene using 5' mismatch primer ATAATGGGTAATATAAAGTGTAATAGAGTA and 3' primer AGAGAACAAACAGACAACATT in a thermal cycler (Eppendorf Hamburg, Germany). PCR cycling conditions comprised of an initial denaturation for 7 min at 95°C followed by 30 repeats (30 sec each) at 95°C, 55°C and 72°C and a final extension step of 7 min at 72°C. Amplified products were verified by electrophoresis using 2% agarose gel. 10 μl aliquots of the amplified products were digested overnight at 37°C with the restriction enzyme Rsa I (Fermentas). Digested products were resolved on 4% agarose gel and cleavage products were visualized by ethidium bromide staining. Deletion genotype was not amenable to restriction digestion and resolved as a single 114 bp band whereas the Insertion genotype was digested by Rsa I enzyme yielding 90 bp and 29 bp fragments. Heterozygous Insertion/Deletion genotype was characterized by all three bands of 114 bp, 90 bp and 29 bp. Results were further validated by direct sequencing. Purification and Sequencing was done by Macrogen Inc.1001 World Meridian Venture Center. The genotypic results were in conformity with the sequencing analysis.
Statistical analyses
Data were managed by Microsoft excel and Java Stat software. Power calculation of the study was performed by online statistical calculator (http://osse.bii.a-star.edu.sg/calculation2.php). The power of study measures 81.4% for observed frequency of minor alleles at 5% significance level. Statistical Package for Social Sciences (SPSS, version 20.0) was used for statistical analysis. Data was described as Mean ± SD and percentage. Chi-square (χ2) test and odds ratio analysis were used for genotyping where P value of <0.05 was considered to be significant. Hardy and Weinberg equilibrium was determined by χ2 analysis. Inter group variants of metric data was measured by Students t-test at 95% confidence interval. The non-metric data was analyzed by Mann Whitney-U test.
Results
-
320
unrelated subjects with type 2 diabetes mellitus and 350 healthy controls were screened for the Del/Ins polymorphism in 3'-UTR of the LEPR gene. The predominant age group was 50–60 years and the mean age of cases and controls was 50.4 ± 11.1 years and 49.2 ± 12.4 years respectively. 69.5% of cases had a familial history of type 2 diabetes. 55.2% cases were shown to be hypertensive by systemic examination. The status of disease was indicated by HbA1c levels. HbA1c > 8% corresponded to 41% of cases, illustrating uncontrolled diabetes state and improper management of disease in such subjects. Molecular characterization of 3'-UTR of the LEPR gene by PCR-RFLP revealed three genotypes (Del/Del, Del/Ins and Ins/Ins) in our population (Figures 1, 2 and 3). Genotypes were in Hardy Weinberg equilibrium for both cases (χ2 = 3.03, P = Non Sig) and controls (χ2 = 2.16, P = Non Sig). The homozygous Del/Del genotype was present in 55.6% of cases and 41.7% of controls. 40.4% of cases and 48.6% of controls exhibited the Del/Ins genotype (Table 1). The rare Ins/Ins genotype was present in only 9.7% of controls and 4% of cases. The homozygous Del genotype was significantly higher in the cases than in controls (P = 0.003). Frequency of Del allele was 0.75and 0.66, while as the frequency of Ins allele was 0.24 and 0.34 in cases and controls respectively (OR: 0.62, 95% CI: 0.48-0.78). The frequency of Del allele correlated directly with the risk factors and the severity of disease as shown by the intra-group genotype analysis (Table 2). Among the cases, there was a significant association between the presence of Del allele and various risk factors. Del allele was observed to be directly associated with elevated BMI, WHR and total cholesterol. Although fasting and post-prandial glucose levels didn’t vary significantly across Del and Ins genotypes, association of HbA1c levels were statistically significant. This can be due to disparity in the glycemic variability or underlying poor control over longer duration. The glycemic control of the studied population is a major reason for extensive difference between plasma glucose and HbA1c levels. Systemic examination of subjects showed that the carriers of Del allele were primarily hypertensive. Higher incidence of Ins allele in the control population is reminiscent of its protective role against the risk factors of T2DM in our population. The intra-group genotyping analysis in controls showed a linear association of the risk factors with the three genotypes: Del/Del, Del/Ins and Ins/Ins. Del allele was associated with atypical profile of various risk factors that play a role in predisposing the carriers of Del allele to T2DM whereas the Ins allele exhibited classic values for the same in the control population.
Discussion
The risk of developing diabetes and related metabolic abnormalities is high in Asian Indians as compared to other ethnic groups and is contributed in part by specific genetic factors determining body fat distribution and glucose metabolism. Distinctive features of “Asian Indian phenotype” include major risk factors responsible for increased predilection towards type 2 diabetes [23]–[27]. Increased level of adipokines that promote insulin resistance is also an underlying biochemical risk factor [28]. The variance in prevalence rates for different ethnic populations substantiates the evidence for a genetic component involved in the development of T2DM. However, non-genetic factors such as environmental and cultural influences can be moderately ascribed to ethnic variability. Studies have shown that while some genes seem to confer increased susceptibility to diabetes in Indians [29],[30]; some protective genes in Europeans do not appear to protect Indians [31]. Kashmir valley is an ethnic region situated in the north India. Very few studies highlighting the prevalence of type 2 diabetes in this region have been undertaken [32],[33]. The population also exhibits high frequency of undiagnosed diabetes [32]. Lately an increased trend in the rate of T2DM along with a high prevalence of impaired fasting glycaemia (IFG) especially in young adults was observed [33]. Although the socio-geographic features of this population like being a distinct, cut-off valley from other states and highly endogamous and consanguineous marriages practised here, the place offers an interesting and potential population for conducting genetic studies. However, studies exploring the genetic variations or molecular mechanisms involved in the etiopathogenesis of type 2 diabetes are even meagre for this population [34],[35].
The foremost advances linking the genetic basis of T2DM has amassed around 40 predisposing loci [3]. A number of sequence determinants are known to affect mammalian mRNA abundance [36]. Occurrence of multiple copies of the core sequence UUAUUUA(U/A)(U/A), designated as the A + U destabilizing element (AUDE), is known to destabilize a variety of mRNA species [37]. A closely reminiscent sequence of 5'-UACUUUAUA-3' is created in the 3'-UTR of the insertion allele. The presence of extra C in this sequence could probably eliminate the destabilizing effect of the core sequence, nonetheless its role is not identified [17]. The 3'-UTR of LEPR gene harbors a pentanucleotide Del/Ins polymorphism that produces an AU-rich sequence capable of affecting the mRNA stability by creating a stem-loop structure [22]. The mechanisms by which stem-loops affect mRNA stability is not completely understood though in many cases these motifs have been shown to bind to regulatory proteins, which in turn could affect the rate of degradation and/or translation of mRNA [36],[38].
We undertook this study to elucidate the role of CTTTA pentanucleotide Del/Ins polymorphism in LEPR gene in ethnic Kashmiri population. In our population frequency of Del allele was 75.78% and 66% and frequency of Ins allele was 24.21% and 34% in cases and controls respectively. The genotypes for this polymorphism were in Hardy Weinberg equilibrium for both cases and controls. In our study, Del allele showed a linear association with the prevalence of diabetes as well as with the associated risk factors. According to our observations, Ins allele conferred a protective effect against the development of T2DM. The genotype and allele frequency of this polymorphism in our population is comparable to another study in which the carriers of the insertion allele had a 79% reduced risk of T2DM compared with non-carriers in the 4-year follow-up [19]. The Del/Ins variant has been found to be associated with serum insulin levels [17],[19], serum high density lipoprotein (HDL)-cholesterol and apolipoprotein A (apo A)-I levels [20], and susceptibility to T2DM [20]–[22]. In our study the intra group genotyping among both cases and controls explicated a positive correlation between the Del allele and the imperative risk factors that predispose our population to T2DM. Our study attests the salvage role of the Ins allele against the predilection and occurrence of T2DM in our population. The elusive factors of relatively higher proportion of IFG in Kashmiri population [32],[33] and the overweight-obesity endemic [39] can in part be explained at the molecular level by the prevalence of a genotypic risk factor (Del allele) that might play a role in inflection and predisposition of its carriers to the T2DM. However other genetic factors may modulate this effect in cumulative or independent manner. The elucidation of these factors in larger samples from our population may substantiate the role of pertinent molecular and genetic factors involved in the pathogenesis and progression to T2DM. This study and its findings may be useful and provide some basic information regarding our understanding of multifaceted pathological interconnections between metabolic legacy, genetic predisposition and contemporary surroundings.
References
Nolan CJ, Damm P, Prentki M: Type 2 diabetes across generations: from pathophysiology to prevention and management. Lancet 2011, 378: 169–181. 10.1016/S0140-6736(11)60614-4
Pinney SE, Simmons RA: Epigenetic mechanisms in the development of type 2 diabetes. Trends Endocrinol Metabol 2010, 21: 223–229. 10.1016/j.tem.2009.10.002
Lyssenko V, Groop L: Genome-wide association study for type 2 diabetes:clinical applications. Curr Opin Lipidol 2009, 20: 87–91. 10.1097/MOL.0b013e32832923af
Florez JC: The genetics of type 2 diabetes: a realistic appraisal in 2008. J Clin Endocrinol Metabol 2008, 93: 4633–4642. 10.1210/jc.2008-1345
Kwon H, Pessin JE: Adipokines mediate inflammation and insulin resistance. Front Endocrinol (Lausanne) 2013, 4: 71.
Tartaglia LA: The leptin receptor. J Biol Chem 1997, 272: 6093–6096. 10.1074/jbc.272.10.6093
Kronenberg HM, Melmed S, Larsen PR, Polonsky KS: Williams Textbook of Endocrinology. Elsevier Health Sciences, Philadelphia, PA; 2008.
Kieffer TJ, Heller SR, Leech CA, Holz GG, Habener JF: Leptin suppression of insulin secretion by activation of ATP-sensitive K channels in pancreatic beta-cells. Diabetes 1997, 46: 1087–1093. 10.2337/diab.46.6.1087
Hummel KP, Dickie MM, Coleman D: Diabetes, a new mutation in the mouse. Science 1996, 153: 1127–1128. 10.1126/science.153.3740.1127
Gullicksen PS, Della-Fera MA, Baile CA: Leptin-induced adipose apoptosis: Implications for body weight regulation. Apoptosis 2003, 8: 327–335. 10.1023/A:1024112716024
Seufert J: Leptin effects on pancreatic beta-cell gene expression and function. Diabetes 2004, 53: 152–158. 10.2337/diabetes.53.2007.S152
Kieffer TJ, Heller RS, Haberner JF: Leptin receptors expressed on pancreatic b-cells. Biochem Biophys Res Commun 1996, 224: 522–527. 10.1006/bbrc.1996.1059
Emilsson V, Liu YL, Cawthorne MA, Morton NM, Davenport M: Expression of the leptin receptor mRNA in pancreatic islets and direct inhibitory action of leptin on insulin secretion. Diabetes 1997, 46: 313–316. 10.2337/diab.46.2.313
Thompson DB, Janssen RC, Ossowski VM, Prochazka M, Knowler WC, Bogardus C: Evidence for linkage between a region on chromosome 1 and the acute insulin response in Pima Indians. Diabetes 1995, 44: 478–481. 10.2337/diab.44.4.478
Thompson DB, Sutherland J, Appel W, Ossowski V: A physical map at 1p31 encompassing the acute insulin response locus and the leptin receptor. Genomics 1997, 39: 227–230. 10.1006/geno.1996.4504
Winick JD, Stoffel M, Friedman JF: Identification of microsatellites linked to the human leptin receptor gene on chromosome 1. Genomics 1996, 36: 221–222. 10.1006/geno.1996.0455
Oksanen L, Kaprio J, Mustajoki P, Kontula K: A common pentanucleotide polymorphism of the 3'-untranslated part of the leptin receptor gene generates a putative stem-loop motif in the mRNA and is associated with serum insulin levels in obese individuals. Int J Obes Relat Metab Disord 1998, 22: 634–640. 10.1038/sj.ijo.0800639
Francke S, Clement K, Dina C, Inoue H, Behn P, Vatin V, Basdevant A, Guy-Grand B, Permutt MA, Froguel P, Hager J: Genetic studies of the leptin receptor gene in morbidly obese French Caucasian families. Hum Genet 1997, 100: 491–496. 10.1007/s004390050540
Lakka HM, Oksanen L, Tuomainen TP, Kontula K, Salonen JT: The common pentanucleotide polymorphism of the 3'-untranslated region of the leptin receptor gene is associated with serum insulin levels and the risk of type 2 diabetes in non-diabetic men: a prospective case–control study. J Intern Med 2000, 248: 77–83. 10.1046/j.1365-2796.2000.00696.x
Nishikai K, Hirose H, Ishii T, Hayashi M, Saito I, Saruta T: Effects of leptin receptor gene 3'-untranslated region polymorphism on metabolic profiles in young Japanese men. J Atheroscler Thromb 2004, 11: 73–78. 10.5551/jat.11.73
Nannipieri M, Posadas R, Bonotti A, Bonotti A, Williams K, Gonzalez-Villalpando C, Stern MP, Ferrannini E: Polymorphism of the 3'-untranslated region of the leptin receptor gene, but not the adiponectin SNP45 polymorphism, predicts type 2 diabetes: a population-based study. Diabetes Care 2006, 29: 2509–2511. 10.2337/dc06-0355
Salopuro T, Pulkkinen L, Lindstrom J, Eriksson JG, Valle TT, Hämäläinen H, Ilanne-Parikka P, Keinänen-Kiukaanniemi S, Tuomilehto J, Laakso M, Uusitupa M: Genetic variation in leptin receptor gene is associated with type 2 diabetes and body weight: the finish diabetes prevention study. Int J Obes (Lond) 2005, 29: 1245–1251. 10.1038/sj.ijo.0803024
Mohan V, Sharp PS, Cloke HR, Burrin JM, Schumer B, Kohner EM: Serum immunoreactive insulin responses to a glucose load in Asian Indian and European Type 2 (noninsulin-dependent) diabetic patients and control subjects. Diabetologia 1986, 29: 235–237. 10.1007/BF00454882
McKeigue PM, Shah B, Marmot MG: Relation of central obesity and insulin resistance with high diabetes prevalence and cardiovascular risk in South Asians. Lancet 1991, 337: 382–386. 10.1016/0140-6736(91)91164-P
Abate N, Chandalia M: Ethnicity and type 2 diabetes: focus on Asian Indians. J Diabetes Complications 2001, 15: 320–327. 10.1016/S1056-8727(01)00161-1
Joshi R: Metabolic syndrome - Emerging clusters of the Indian phenotype. J Assoc Physicians India 2003, 51: 445–446.
Mohan V, Sandeep S, Deepa R, Shah B, Varghese C: Epidemiology of type 2 diabetes: Indian scenario. Indian J Med Res 2007, 125: 217–230.
Abate N, Chandalia M, Snell PG, Grundy SM: Adipose tissue metabolites and insulin resistance in nondiabetic Asian Indian men. J Clin Endocrinol Metab 2004, 89: 2750–2755. 10.1210/jc.2003-031843
Abate N, Chandalia M, Satija P, Adams-Huet B, Grundy SM, Sandeep S, Radha V, Deepa R, Mohan V: ENPP1/PC-1 K121Q polymorphism and genetic susceptibility to type 2 diabetes. Diabetes 2005, 54: 1207–1213. 10.2337/diabetes.54.4.1207
Vimaleswaran KS, Radha V, Ghosh S, Majumder PP, Deepa R, Babu HN: Peroxisome proliferator-activated receptor-gamma co-activator-1alpha (PGC-1alpha) gene polymorphisms and their relationship to Type 2 diabetes in Asian Indians. Diabet Med 2005, 22: 1516–1521. 10.1111/j.1464-5491.2005.01709.x
Radha V, Vimaleswaran KS, Babu HN, Abate N, Chandalia M, Satija P: Role of genetic polymorphism peroxisome proliferator-activated receptor-gamma2 Pro12Ala on ethnic susceptibility to diabetes in South-Asian and Caucasian subjects: Evidence for heterogeneity. Diabetes Care 2006, 29: 1046–1051. 10.2337/dc05-1473
Zargar AH, Khan AK, Masoodi SR, Laway BA, Wani AI, Bashir MI, Dar FA: Prevalence of type 2 diabetes mellitus and impaired glucose tolerance in the Kashmir Valley of the Indian subcontinent. Diab Res Clin Pract 2000, 47: 135–146. 10.1016/S0168-8227(99)00110-2
Zargar AH, Wani AA, Laway BA, Masoodi SR, Wani AI, Bashir MI, Dar FA: Prevalence of diabetes mellitus and other abnormalities of glucose tolerance in young adults aged 20–40 years in North India (Kashmir Valley). Diab Res Clin Pract 2008, 82: 276–281. 10.1016/j.diabres.2008.08.006
Hameed I, Afroze D, Masoodi SR, Bhat RA, Ganai BA: Screening Codon 233, 234 and 276 Polymorphisms in Exon 3 of Insulin Receptor Gene in Type 2 Diabetes Mellitus Patients of Kashmir Valley. IOSR J Pharm Biol Sci 2012, 3: 7–10.
Hameed I, Masoodi SR, Afroze D, Naykoo NA, Bhat RA, Ganai BA: Trp homozygotes at codon 64 of ADRB3 gene are protected against the risk of type 2 diabetes in the Kashmiri population. Genet Test Mol Biomarkers 2013, 17: 775–779. 10.1089/gtmb.2013.0297
Ross J: mRNA stability in mammalian cells. Microbiol Rev 1995, 59: 423–450.
Brown CY, Lagnado CA, Goodall GJ: A cytokine mRNA destabilizing element that is structurally and functionally distinct from AU-rich elements. Proc Natl Acad Sci U S A 1996, 93: 13721–13725. 10.1073/pnas.93.24.13721
Wang Z, Kiledjian M: The poly (A)-binding protein and an mrna stability protein jointly regulate an endoribonuclease activity. Mol Cell Biol 2000, 20: 6334–6341. 10.1128/MCB.20.17.6334-6341.2000
Masoodi SR, Wani AA, Wani AI, Bashir MI, Laway BA, Zargar AH: Prevalence of overweight and obesity in young adults aged 20–40 years in North India (Kashmir Valley). Diabetes Res Clin Pract 2010, 87: 4–6. 10.1016/j.diabres.2009.11.004
Acknowledgement
The authors are thankful to Department of Science and Technology, Government of India for providing funding under Women Scientist Scheme (DST WOSA: LS 509/2012).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
IH, BAG and SRM conceived and designed the study. DA, RAB aided in acquiring data and drafting. NAN, SAM and IM analysed and interpreted data. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/4.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly credited. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hameed, I., Masoodi, S.R., Afroze, D. et al. CTTTA Deletion/Insertion polymorphism in 3'-UTR of LEPR gene in type 2 diabetes subjects belonging to Kashmiri population. J Diabetes Metab Disord 13, 124 (2014). https://doi.org/10.1186/s40200-014-0124-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s40200-014-0124-z