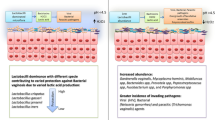
Abstract
Chlamydia trachomatis, Mycoplasma spp., Neisseria gonorrhoeae and Treponema pallidum are sexually transmitted pathogens that threaten reproductive health worldwide. In this study, vaginal swabs obtained from women (n = 133) that attended an infertility clinic in China were tested with qPCRs for C. trachomatis, Mycoplasma spp., N. gonorrhoeae, T. pallidum and tetracycline resistance genes. While none of vaginal swabs were positive for N. gonorrhoeae and T. pallidum, 18.8% (25/133) of the swabs were positive for Chlamydia spp. and 17.3% of the swabs (23/133) were positive for Mycoplasma species. All swabs tested were positive for tetracycline resistance gene tet(M) which is the most effective antibiotic for bacterial sexually transmitted infections. The qPCRs determined that the gene copy number per swab for tet(M) was 7.6 times as high as that of C. trachomatis 23S rRNA, and 14.7 times of Mycoplasma spp. 16S rRNA. In China, most hospitals do not detect C. trachomatis and Mycoplasma spp. in women with sexually transmitted infections and fertility problems. This study strongly suggests that C. trachomatis and Mycoplasma spp. should be routinely tested in women with sexually transmitted infections and infertility in China, and that antimicrobial resistance of these organisms should be monitored. Further studies are warranted to determine the prevalences in different regions and associated risk factors.
Similar content being viewed by others
Introduction
Female infertility, including vaginal multi-pathogen infection induced infertility, is a major public health concern worldwide. Chlamydia trachomatis, Mycoplasma spp., Neisseria gonorrhoeae and Treponema pallidum have been extensively shown to be associated with infertility, particularly because of endometrial and tubal inflammation.
Chlamydia trachomatis is an obligate intracellular bacterial pathogen which remains the leading cause of bacterial sexually transmitted disease worldwide. Infections in the lower genital tract are frequently asymptomatic and, if untreated, can ascend to the upper genital tract, potentially leading to complications such as tubal factor infertility, and subfertility (Tang et al. 2015; Karinen et al. 2004). The organism is often undiagnosed in routine examinations performed in infertility clinical in China (Zheng et al. 2017).
Mycoplasmas are frequently isolated from the genital tract with M. genitalium and M. hominis considered responsible for genital diseases, infertility, and obstetric complications (Haggerty and Ness 2007; Taylor-Robinson and Lamont 2011; Kataoka et al. 2006). M. hominis has been found in about two-thirds of women with bacterial vaginosis and in 10% of women with salpingitis and endometritis (Judlin 2003).
Neisseria gonorrhoeae is the etiological agent of gonorrhea, the second most frequently reported sexually transmitted infection (STI) in the world. N. gonorrhoeae is a cause of pelvic inflammatory disease in women, which can lead to serious reproductive complications including tubal infertility, ectopic pregnancy, and chronic pelvic pain (Costa Lourenço et al. 2017; Kirkcaldy et al. 2016). The World Health Organization reported over 78 million new cases of gonococcal infection in people aged 15–49 worldwide during 2012 (World Health Organization 2016). Gonorrhea is usually symptomatic in men. However, there can be symptomatic gonococcal cervicitis and the complicated gonorrhea may cause infertility in women (Costa Lourenço et al. 2017; Ison 2011).
Treponema pallidum can lead to the complex and systemic disease, syphilis which reduces the clinical pregnancy rate after in vitro fertilization/intra-cytoplasmic sperm injection (Wang et al. 2015). Furthermore, T. pallidum infection with varied clinical presentations can cause neurological, cardiovascular and other multisystem damage, leading to a long time course and serious, even life-threatening consequences.
Tetracycline is the antimicrobial of choice against bacterial STIs including C. trachomatis, Mycoplasmas and Ureaplasma. There is a high-level of resistance to tetracyclines in genital bacteria including lactobacilli which is due mainly to the presence of the tet(M) gene that mediates resistance to tetracyclines (de Barbeyrac et al. 1996; Dégrange et al. 2008; Mardassi et al. 2012).
The aim of the present work was to evaluate the presence of C. trachomatis, Mycoplasma spp., N. gonorrhoeae, T. pallidum and tetracycline resistance gene in vaginal swabs of women undergoing an infertility evaluation in China.
Materials and methods
Vaginal samples
The protocols used in the study were approved by the Institutional Review Board of Northern Jiangsu People’s Hospital of China. In September of 2015, vaginal swabs were obtained from 133 women undergoing infertility evaluation in an infertility clinic of Northern Jiangsu Peoples Hospital, Yangzhou city, Jiangsu Province of China. Swabs were collected into sterile tubes containing 400 μl DNA/RNA stabilization buffer (Roche Molecular Biochemicals, Indianapolis, IN, USA) and stored at − 80 °C until the DNA was extracted. In addition, a questionnaire that surveyed demographic details as well as obstetric and gynecologic history was administered.
DNA extraction
The High-Pure PCR Template Preparation Kit (Roche Molecular Biochemicals, Indianapolis, IN, USA) was used to extract total nucleic acids from vaginal swabs according to the manufacturer’s instructions and as described before (Li et al. 2016). The extracted DNA was eluted in 200 μl elution buffer.
PCR assays
Chlamydia FRET-qPCR
The FRET-qPCR used in this study followed the protocols described by DeGraves et al. (2003) and Guo et al. (2016) and was performed in a LightCycler 480-II real-time PCR platform. This PCR assay targets 168-bp fragment of the Chlamydia spp. 23S rRNA gene, and could detect all 11 Chlamydia species with a detection sensitivity of single copy/reaction. The PCR products were further verified by electrophoresis through 2% agarose gels (BIOWEST®, Hong Kong, China), purification using the QIAquick PCR Purification Kit (Qiagen), and sequencing with forward and reverse primers (BGI, Shanghai, China).
Mycoplasma FRET-qPCR
To investigate the presence of Mycoplasma in the vaginal swabs, a set of primers and probes were designed using Vector NTI to amplify a 174-bp fragment of the Mycoplasma spp. 16S rRNA gene (Table 1). PCR amplification was performed in a LightCycler 480-II real-time PCR platform using a high-stringency 18-cycle step-down temperature protocol: 6 × 1 s @ 95 °C, 12 s @64 °C, 8 s @ 72 °C; 9 × 1 s @ 95 °C, 12 s @ 62 °C, 8 s @ 72 °C; 3 × 1 s @ 95 °C, 12 s @ 60 °C, 8 s @ 72 °C; followed by 30 low-stringency cycles: 30 × 1 s @ 95 °C, 12 s @ 57 °C, 30 s @ 67 °C, and 10 s @ 72 °C. Twenty μl PCR reactions were prepared containing 10.0 μl DNA template, 0.2 μl forward primer (100 μM), 0.2 μl reverse primer (100 μM,), 4.0 μl 5× PCR buffer, 0.4 μl 10 μM dNTP, 0.3 μl 5 U/μl Taq DNA polymerase and 4.9 μl ultrapure H2O. The PCR products were further verified, purified, and sequenced as mentioned above. A standard PCR with a long amplicon (703–713 bp) of the 16S rRNA gene were performed on positive samples based on FRET-PCR to determine the Mycoplasma species (Yoshida et al. 2002).
For use as quantitative standards, the products of the Mycoplasma FRET-PCR on vaginal swabs were gel purified using a QIAquick Gel Extraction Kit (Qiagen, Valencia, CA, USA). An aliquot of the purified product was sequenced for confirmation at GenScript (Nanjing Jiangsu, China) and the remainder quantified (ng/ml) with the Quanti-iT™ PicoGreen® dsDNA Assay Kit. After using the molecular mass of the 16S rRNA gene to calculate the molarity of the solution, dilutions were made to give solutions containing 10,000, 1000, 100, 10, and 1 gene copies per reaction. These were amplified by Mycoplasma FRET-PCR in triplicate to determine the detection limit of the PCR.
The specificity of this PCR was further verified with the amplification of DNAs s from Salmonella Typhimurium (ATCC 14028), Escherichia coli (ATCC25922), C. trachomatis (ATCC VR-571B), Ehrlichia canis, Anaplasma phagocytophilum and Rickettsia felis.
Neisseria gonorrhoeae and T. pallidum qPCR
Two sets of primers were used to detect N. gonorrhoeae and T. pallidum by the porA pseudogene and the polA gene respectively as described (Whiley et al. 2004; Heymans et al. 2010).
RPPs gene qPCR
A qPCR was established to quantify a class of tetracycline resistance genes encoding for ribosomal protection proteins (RPPs) in vaginal swabs in this study. The tet(M), tet(S), tet(O), tet(Q), tet(T), tet(36), tet(44) sequences were obtained from NCBI (http://www.ncbi.nlm.nih.gov). Using the Clustal Multiple Alignment Algorithm, we identified a highly conserved 245-bp PCR target on the above seven tetracycline resistance genes (Table 1). The thermal protocol was identical to what described above for the Mycoplasma-qPCR. The specificity of the primers was verified by BLASTn and DNA sequencing of obtained PCR products. The sensitivity of the tet(M)-qPCR was determined as mentioned above.
Statistical analysis
All statistical analyses were performed with the Statistica 7.0 software package (StatSoft, Inc., Oklahoma, USA). Positivity of pathogens in different age groups was compared using the Chi squared Test. Differences of copy numbers of pathogens and tet(M) gene in different age groups were logarithmically transformed and analyzed with the two-tailed Tukey honest significant difference (HSD) test in one-way ANOVA. Differences at P ≤ 0.05 were considered significant.
Results
Establishment of the Mycoplasma FRET qPCR and tet(M) qPCR
The Mycoplasma FRET qPCR established in this study detected the target Mycoplasma 16S rRNA with a detection limit of one copy 16S rRNA per reaction. It did not detect Salmonella Typhimurium, Escherichia coli, C. trachomatis, Ehrlichia canis, Anaplasma phagocytophilum and Rickettsia felis. The RPPs gene-qPCR we established had a detection limit of one gene copy per reaction while its specificity was verified by sequencing.
Prevalence and copy numbers of C. trachomatis, Mycoplasma spp., N. gonorrhoeae, T. pallidum and tet(M) in vaginal swabs from infertile women
While none of the swabs were positive for N. gonorrhoeae and T. pallidum, 31.6% of the swabs (42/133) were positive for C. trachomatis and/or Mycoplasmas infection. In six women (4.5%) coinfection with both C. trachomatis and Mycoplasma spp. was observed. All the swabs were positive for the tet(M) gene.
C. trachomatis was found to be the only chlamydial species in all vaginal swabs with a positivity of 18.8% (25/133). The average gene copy number of C. trachomatis was 27,855 (± 21,301 SEM) per swab.
The FRET-qPCR followed by standard PCR that generates a longer amplicon of the 16S rRNA gene determined that 17.3% (23/133) of swabs were positive for Mycoplasma spp. Sequence analysis of these amplicons revealed that M. spermatophilum (MF769616, n = 2; MF769617, n = 2), M. hominis (MF769618, n = 14), M. faucium (MF769619, n = 4), and Candidatus Mycoplasma girerdii (Ca. M. girerdii) (MF769620, n = 1) (Fig. 1). The average gene copy number for Mycoplasma spp. was 14,433 (± 7872 SEM) per swab.
Neighbor-joining phylogenetic tree based on the sequence alignment of the 16S rRNA gene (700 bp). The five strains identified in our study (MF769616–MF769620) are in red font. The sequence identified in our study (MF769616 and MF769617) are most similar to M. spermatophilum, MF769618 is most similar to M. hominis, MF769619 is most similar to M. faucium, MF769620 is most similar to Ca. M. girerdii. Bootstrap percentage values greater than 50% are given at the nodes of the tree (1000 replicates)
Sequence analysis of 22 RPPs PCR products revealed eight different tet(M) sequences (MF769608–MF769615) (Fig. 2). The sequences of MF769608 and MF769620 in this study were identical to the tet(M) sequences (KU545550 and MF422120) in GenBank.
Neighbor-joining phylogenetic tree based on sequences of the RPPs gene (245 bp). The tetracycline resistance genes identified in our study (MF769608–MF769615, in red font) are the most similar to tet(M). Bootstrap percentage values greater than 50% are given at the nodes of the tree (1000 replicates)
All the vaginal swabs were positive for tet(M) with the average gene copy number per swabs being 212,878 (± 39,025 SEM) which was 7.6 times that for C. trachomatis 23S rRNA and 14.7 times that for Mycoplasma spp. 16S rRNA.
Prevalence and copy numbers of C. trachomatis, Mycoplasma and tet(M) in different age groups
The average age of the 128 women for whom we had data was 29.35 years (range 21–44 years). For analysis we divided the women into four groups: 21–25, 26–31, 32–37, and 38–44 years.
The positivity for C. trachomatis in the 38–44 age group (42.9%) was significantly higher (P < 0.01) than in the younger age groups (32–37 years, 25.0%; 21–25 age group (21.4%) and 26–31 age group (15.7%) (Fig. 3). Similarly, the positivity for Mycoplasma in the 21–25 age group (7.1%) was significant lower (P < 0.01) than in the older age groups (26–31 age group, 20.5%; 32–37 age group, 16.7%; 38–44 age group, 14.3%) (Fig. 3).
Prevalence and copy numbers of C. trachomatis, Mycoplasma and tet(M) gene in vaginal samples in four age groups. a The C. trachomatis positivity in 21–25 age group, 26–31 age group, 32–37 age group and 38–44 age group detected in this study were 21.4, 15.7, 25.0 and 42.9% respectively. The Mycoplasma positivity in above four age groups detected in this study were 7.1, 20.5, 16.7 and 14.3% respectively. The tet(M) gene positivity in above four age groups were all 100%. b The average gene copy numbers for C. trachomatis 23S rRNA, Mycoplasma spp. 16S rRNA and tet(M) gene were 102.53 ± 1.05 SEM, 102.81 ± 1.24 SEM, 104.28 ± 1.34 SEM (being equivalent to 27,855 ± 21,301 SEM; 14,433 ± 7872 SEM; 212,878 ± 39,025 SEM), respectively. The gene copy number for four age groups did not differ significantly for C. trachomatis, Mycoplasma spp. and tet(M). However, the tet(M) copy number is significantly higher than C. trachomatis (averagely 7.6 times), and Mycoplasma spp. (averagely 14.7 times)
The average gene copy numbers of C. trachomatis, Mycoplasma and tet(M) did not differ significantly between the four age groups (Fig. 3) although the total gene copy number per swab was significantly higher for tet(M) (P < 10−4) than for C. trachomatis 23S rRNA and Mycoplasma spp. 16S rRNA.
Discussion
This study demonstrated that 31.6% of women attending an infertility clinic in China tested positive for C. trachomatis and/or Mycoplasma spp. whereas these two pathogens are usually not detected in women with STIs and fertility problems in most Chinese hospitals (Peng et al. 2017; Bao et al. 2016). In contrast, we failed to identify the two pathogens (N. gonorrhoeae and T. pallidum) that are at the top of the list for STI surveillance in China. Surprisingly, all the swabs tested in this study were positive for tet(M), the resistance gene against tetracycline which is the most commonly used and most effective antimicrobial for bacterial STIs. Our study strongly suggests that C. trachomatis and Mycoplasma spp. should be routinely tested for in women with STIs and infertility in China, and that the antimicrobial resistance of these organisms should be monitored.
The Chinese government launched a massive campaign to eliminate STIs in the 1950s, and STIs were thought to be extremely uncommon by the 1960s in China (Cohen et al. 1997). However, since the early 1980s, STIs have reemerged with the introduction of the open door policy and economic liberalization and are now recognized as a major public-health problem in China as they have spread from the high risk group to the general population (Chen et al. 2007).
In China, HIV/AIDS, syphilis, and gonorrhea are reportable STIs according to the Law of the People’s Republic of China on Prevention and Treatment of Infectious Diseases. The reported incidence of primary and secondary syphilis was 11.7 cases per 100,000 residents in 2009, an increase of 2.1-fold since 2005 (Chen et al. 2011). However, the reported incidence of gonorrhea has decreased by about 30% (Chen et al. 2011) with the overall prevalence of N. gonorrhoeae infection in the general population being 0.08% for women and 0.02% for men (Parish et al. 2003). The overall trend is that the prevalence of N. gonorrhoeae and T. pallidum has dropped significantly in China (Chen et al. 2011; Adachi et al. 2016), congruent to the findings as none of the 133 vaginal swabs were tested positive for N. gonorrhoeae and T. pallidum in this study.
C. trachomatis infections of the lower female genital tract are frequently asymptomatic. However, if infections do not resolve or persist untreated, organisms can ascend and pathology in the upper genital tract, potentially causing salpingitis and functional damage to the fallopian tubes and tubal factor infertility (Hafner 2015). The incidence of genital C. trachomatis infections increased by nearly 40% from 37.20 per 100,000 persons in 2010 and 51.3 per 100,000 in 2014 in Guangdong Province of China (Wong et al. 2017). Based on a Chinese Health and Family Life Survey, the prevalence of C. trachomatis was 2.6% (95% CI 1.6–4.1%) in Chinese women (Conejero et al. 2013; Parish et al. 2003). This is similar to the 2.7% (n = 1717) positivity for women reported in the Netherlands (Morré et al. 1999). While the international standards recommend annual screening for C. trachomatis in sexually active women (Conejero et al. 2013), most hospitals in China do not detect C. trachomatis in women during regular checks or in women with STIs and fertility problems. The high prevalence of C. trachomatis demonstrated in this study strongly suggests that hospitals in China should follow the international recommendation and national guideline for routine surveillance of C. trachomatis.
Mycoplasma species have been reported to be associated with perinatal morbidity and mortality while M. hominis has been mostly associated with chorioamnionitis and thought to be an etiological agent (Taylor-Robinson and Lamont 2011). The overall prevalence of urogenital Mycoplasma infections varies in different countries while international reports suggest an increase in infections due to Mycoplasma over the last decade (Díaz et al. 2013). The global prevalence of M. genitalium among symptomatic and asymptomatic sexually active women ranges between 1 and 6.4% (Pereboom et al. 2014). This variability in prevalence rates reported in different countries is perhaps due to differences in detection methods, types of samples studied, sample sizes, hygiene issues, socioeconomic status, age of participants, and absence of regular screening, treatment, and control programs (Ahmadi et al. 2016). This study identified 22/23 of Mycoplasmas sequences being hominis taxa, and only one sequence was identified as Ca. M. girerdii.
For decades, tetracyclines, as broad-spectrum antibiotics, have been used extensively for treating bacteria-induced STIs. Resistance to tetracyclines is now increasingly prevalent in STIs, and the drugs usefulness in treating urogenital infections is decreasing because of the presence of the tet(M). Surprisingly, in our study 100% of the swabs we tested were positive for tet(M) gene which was present in very high copy numbers, 7.6 times that of C. trachomatis 23S rRNA and 14.7 times that of Mycoplasma spp. 16S rRNA. This indicates the C. trachomatis and Mycoplasma spp. carried multiple copies of the tet(M) or that the gene was also present in other vaginal microbes such as lactobacilli. The antimicrobial resistance features of vaginal bacteria should be regularly monitored to provide the judicious treatment of STIs. The phylogenetic analysis based on eight sequences identified in this study and 12 species RPPs gene in GenBank revealed that all eight sequences belong to tet(M) taxa, which is one of the most prevalent class of RPPs gene conferring antibiotic resistance.
Traditionally, the diagnosis of genitourinary pathogens has been based on bacterial culture. Bacterial isolation, however, is cumbersome, costly, and time-consuming. It is also selective in that samples must be appropriately collected, transported and stored to maximize the number of viable bacteria. It is particularly difficult to grow obligate intracellular bacterial such as C. trachomatis while cell lines or chicken embryos are required for their growth. In this study, sensitive and specific qPCRs were applied for direct testing of samples for pathogens and resistant genes. This approach avoids the limitations of bacterial isolation and the resultant underestimation of pathogen prevalence.
In conclusion, we found a high prevalence of C. trachomatis, Mycoplasma spp. and tet(M) in vaginal swabs from women with fertility problems in China, whilst N. gonorrhoeae or T. pallidum were not detected. While more studies are warranted to investigate the prevalences of these organisms and tet(M) in larger populations from different regions, our data should alert health workers in China that C. trachomatis and Mycoplasma spp. might be the main pathogens in women with STIs and infertility problem.
Abbreviations
- tet :
-
tetracycline resistance gene
- FRET-qPCR:
-
fluorescence resonance energy transfer quantitative PCR
- STI:
-
sexually transmitted infection
- RPPs:
-
ribosomal protection proteins
References
Adachi K, Nielsen Saines K, Klausner JD (2016) Chlamydia trachomatis infection in pregnancy: the global challenge of preventing adverse pregnancy and infant outcomes in Sub-Saharan Africa and Asia. Biomed Res Int 2016:9315757. https://doi.org/10.1155/2016/9315757
Ahmadi MH, Mirsalehian A, Bahador A (2016) Prevalence of urogenital Mycoplasmas in Iran and their effects on fertility potential: a systematic review and meta-analysis. Iran J Public Health 45(4):409–422
Bao J, Yang YJ, Liu H (2016) Genitourinary Mycoplasma infection and drug resistance in eugenics health examination. Chin J Clin Ration Drug Use 9:96. https://doi.org/10.15887/j.cnki.13-1389/r.2016.30.045
Chen ZQ, Zhang GC, Gong XD, Lin C, Gao X, Liang GJ, Yue XL, Chen XS, Cohen MS (2007) Syphilis in China: results of a national surveillance programme. Lancet 369(9556):132–138
Chen XS, Peeling RW, Yin YP, Mabey DC (2011) The epidemic of sexually transmitted infections in China: implications for control and future perspectives. BMC Med 9:111. https://doi.org/10.1186/1741-7015-9-111
Cohen MS, Hoffman IF, Royce RA, Kazembe P, Dyer JR, Daly CC, Zimba D, Vernazza PL, Maida M, Fiscus SA, Eron JJ Jr (1997) Reduction of concentration of HIV-1 in semen after treatment of urethritis: implications for prevention of sexual transmission of HIV-1. AIDSCAP Malawi Research Group. Lancet 349(9069):1868–1873
Conejero C, Cannoni G, Merino PM, Bollmann J, Hidalgo C, Castro M, Schulin-Zeuthen C (2013) Screening of Neisseria gonorrhoeae and Chlamydia trachomatis using techniques of self collected vaginal sample in young women. Rev Chilena Infectol 30(5):489–493. https://doi.org/10.4067/S0716-10182013000500004
Costa Lourenço APRD, Barros Dos Santos KT, Moreira BM, Fracalanzza SEL, Bonelli RR (2017) Antimicrobial resistance in Neisseria gonorrhoeae: history, molecular mechanisms and epidemiological aspects of an emerging global threat. Braz J Microbiol 48:617–628. https://doi.org/10.1016/j.bjm.2017.06.001
de Barbeyrac B, Dupon M, Rodriguez P, Renaudin H, Bébéar C (1996) A Tn1545-like transposon carries the tet(M) gene in tetracycline resistant strains of Bacteroides ureolyticus as well as Ureaplasma urealyticum but not Neisseria gonorrhoeae. J Antimicrob Chemother 37(2):223–232
Dégrange S, Renaudin H, Charron A, Bébéar C, Bébéar CM (2008) Tetracycline resistance in Ureaplasma spp. and Mycoplasma hominis: prevalence in Bordeaux, France, from 1999 to 2002 and description of two tet(M)-positive isolates of M. hominis susceptible to tetracyclines. Antimicrob Agents Chemother 52(2):742–744
DeGraves FJ, Gao D, Hehnen HR, Schlapp T, Kaltenboeck B (2003) Quantitative detection of Chlamydia psittaci and Chlamydia pecorum by high-sensitivity real-time PCR reveals high prevalence of vaginal infection in cattle. J Clin Microbiol 41(4):1726–1729
Díaz L, Cabrera LE, Fernández T, Ibáñez I, Torres Y, Obregón Y, Rivero Y (2013) Frequency and antimicrobial sensitivity of Ureaplasma urealyticum and Mycoplasma hominis in patients with vaginal discharge. MEDICC Rev 15(4):45–47
Guo W, Li J, Kaltenboeck B, Gong J, Fan W, Wang C (2016) Chlamydia gallinacea, not C. psittaci, is the endemic chlamydial species in chicken (Gallus gallus). Sci Rep 6:19638. https://doi.org/10.1038/srep19638
Hafner LM (2015) Pathogenesis of fallopian tube damage caused by Chlamydia trachomatis infections. Contraception 92(2):108–115. https://doi.org/10.1016/j.contraception.2015.01.004
Haggerty CL, Ness RB (2007) Newest approaches to treatment of pelvic inflammatory disease: a review of recent randomized clinical trials. Clin Infect Dis 44(7):953–960
Heymans R, van der Helm JJ, de Vries HJ, Fennema HS, Coutinho RA, Bruisten SM (2010) Clinical value of Treponema pallidum real-time PCR for diagnosis of syphilis. J Clin Microbiol 48(2):497–502. https://doi.org/10.1128/JCM.00720-09
Ison CA (2011) Biology of Neisseria gonorrhoeae and the clinical picture of infection. In: Gross GE, Tyring SK (eds) Sexually transmitted infections and sexually transmitted diseases. Springer, Berlin, pp 77–90
Judlin P (2003) Genital Mycoplasmas. Gynecol Obstet Fertil 31(11):954–959
Karinen L, Pouta A, Hartikainen AL, Bloigu A, Paldanius M, Leinonen M, Saikku P, Järvelin MR (2004) Association between Chlamydia trachomatis antibodies and subfertility in the Northern Finland Birth Cohort 1966 (NFBC 1966), at the age of 31 years. Epidemiol Infect 132(5):977–984
Kataoka S, Yamada T, Chou K, Nishida R, Morikawa M, Minami M, Yamada H, Sakuragi N, Minakami H (2006) Association between preterm birth and vaginal colonization by Mycoplasmas in early pregnancy. J Clin Microbiol 44(1):51–55
Kirkcaldy RD, Harvey A, Papp JR, Del Rio C, Soge OO, Holmes KK, Hook EW 3rd, Kubin G, Riedel S, Zenilman J, Pettus K, Sanders T, Sharpe S, Torrone E (2016) Neisseria gonorrhoeae antimicrobial susceptibility surveillance—The Gonococcal Isolate Surveillance Project, 27 Sites, United States, 2014. MMWR Surveill Summ 65(7):11–19. https://doi.org/10.15585/mmwr.ss6507a1
Li J, Guo W, Kaltenboeck B, Sachse K, Yang Y, Lu G, Zhang J, Luan L, You J, Huang K, Qiu H, Wang Y, Li M, Yang Z, Wang C (2016) Chlamydia pecorum is the endemic intestinal species in cattle while C. gallinacea, C. psittaci and C. pneumoniae associate with sporadic systemic infection. Vet Microbiol 193:93–99. https://doi.org/10.1016/j.vetmic.2016.08.008
Mardassi BB, Aissani N, Moalla I, Dhahri D, Dridi A, Mlik B (2012) Evidence for the predominance of a single tet(M) gene sequence type in tetracycline-resistant Ureaplasma parvum and Mycoplasma hominis isolates from Tunisian patients. J Med Microbiol 61(Pt 9):1254–1261. https://doi.org/10.1099/jmm.0.044016-0
Morré SA, Van Valkengoed IG, Moes RM, Boeke AJ, Meijer CJ, Van den Brule AJ (1999) Determination of Chlamydia trachomatis prevalence in an asymptomatic screening population: performances of the LCx and COBAS Amplicor tests with urine specimens. J Clin Microbiol 37(10):3092–3096
Parish WL, Laumann EO, Cohen MS, Pan S, Zheng H, Hoffman I, Wang T, Ng KH (2003) Population-based study of chlamydial infection in China: a hidden epidemic. JAMA 289(10):1265–1273
Peng Y, Tang M, Feng SS (2017) Prevalence of Chlamydia trachomatis and Ureaplasma urealyticum in 7089 genitourinary samples from women. Lab Med Clin 14:982
Pereboom MT, Spelten ER, Manniën J, Rours GI, Morré SA, Schellevis FG, Hutton EK (2014) Knowledge and acceptability of Chlamydia trachomatis screening among pregnant women and their partners; a cross-sectional study. BMC Public Health 14:704. https://doi.org/10.1186/1471-2458-14-704
Tang L, Chen J, Zhou Z, Yu P, Yang Z, Zhong G (2015) Chlamydia-secreted protease CPAF degrades host antimicrobial peptides. Microbes Infect 17(6):402–408. https://doi.org/10.1016/j.micinf.2015.02.005
Taylor-Robinson D, Lamont RF (2011) Mycoplasmas in pregnancy. BJOG 118(2):164–174. https://doi.org/10.1111/j.1471-0528.2010.02766.x
Wang J, Zhao X, Yuan P, Fang T, Ouyang N, Li R, Ou S, Wang W (2015) Clinical outcomes of in vitro fertilization among Chinese infertile couples treated for syphilis infection. PLoS ONE 10(7):e0133726. https://doi.org/10.1371/journal.pone.0133726 (eCollection 2015)
Whiley DM, Buda PJ, Bayliss J, Cover L, Bates J, Sloots TP (2004) A new confirmatory Neisseria gonorrhoeae real-time PCR assay targeting the porA pseudogene. Eur J Clin Microbiol Infect Dis 23(9):705–710
WHO. Guidelines for the treatment of Neisseria gonorrhoeae (2016) World Health Organization, Geneva. http://www.ncbi.nlm.nih.gov/books/NBK379221/
Wong WC, Zhao Y, Wong NS, Parish WL, Miu HY, Yang LG, Emch M, Ho KM, Fong FY, Tucker JD (2017) Prevalence and risk factors of chlamydia infection in Hong Kong: a population-based geospatial household survey and testing. PLoS ONE 12(2):e0172561. https://doi.org/10.1371/journal.pone.0172561 (eCollection 2017)
Yoshida T, Maeda S, Deguchi T, Ishiko H (2002) Phylogeny-based rapid identification of Mycoplasmas and Ureaplasmas from urethritis patients. J Clin Microbiol 40(1):105–110. https://doi.org/10.1128/JCM.40.1.105-110.2002
Zheng Y, Shi WD, Hu Q (2017) Epidemiological analysis of genital Chlamydia trachomatis in Wuhan of China (2009–2015). J Public Health Prev Med 28:57–59 (in Chinese)
Authors’ contributions
CW conceived of the study. ML, XZ, KH, HQ, JZ and YK performed experiment, computational analysis. CW and ML wrote the paper. All authors read and approved the final manuscript.
Acknowledgements
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
Sequences are deposited in GenBank with Accession Numbers MF769608–MF769620.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The study was approved by the Institutional Review Board of Northern Jiangsu People’s Hospital of China. All patients provided written consent prior to sample collection.
Funding
This work was supported by a grant from National Key Research Project of China (No: 2016YFD0500804) and a grant from the National Natural Science Foundation of China (No: 31272575), and by the Priority Academic Program Development of Jiangsu Higher Education Institutions, Yangzhou, Jiangsu, P. R. China.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Li, M., Zhang, X., Huang, K. et al. Presence of Chlamydia trachomatis and Mycoplasma spp., but not Neisseria gonorrhoeae and Treponema pallidum, in women undergoing an infertility evaluation: high prevalence of tetracycline resistance gene tet(M). AMB Expr 7, 206 (2017). https://doi.org/10.1186/s13568-017-0510-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13568-017-0510-2