Abstract
Background
The common cockle Cerastoderma edule plays an important ecological role in the marine ecosystem both as an infaunal engineer (reef forming and bioturbation) and a food source for protected bird species in its European range. Cockle beds are found in close proximity to aquaculture and fisheries operations, which can be “hot spots” for infectious agents including viruses and bacteria. Ostreid herpesvirus-1 microVar (OsHV-1 μVar) has spread to many Pacific oyster Crassostrea gigas culture sites globally, where it has been associated with significant mortalities in this cultured bivalve. Knowledge on the impact of the virus on the wider ecosystem, is limited. As the likelihood of released virus dispersing into the wider aquatic ecosystem is high, the plasticity of the virus and the susceptibility of C. edule to act as hosts or carriers is unknown.
Methods
In this study, wild C. edule were sampled biweekly at two C. gigas culture sites over a four-month period during the summer when OsHV-1 μVar prevalence is at its highest in oysters. C. edule were screened for the virus molecularly (PCR, qPCR and Sanger sequencing) and visually (in situ hybridisation (ISH)). The cockle’s ability to act as a carrier and transmit OsHV-1 μVar to the oyster host at a temperature of 14 ℃, when the virus is considered to be dormant until water temperatures exceed 16 ℃, was also assessed in laboratory transmission trials.
Results
The results demonstrated that OsHV-1 μVar was detected in all C. edule size/age cohorts, at both culture sites. In the laboratory, viral transmission was effected from cockles to naïve oysters for the first time, five days post-exposure. The laboratory study also demonstrated that OsHV-1 μVar was active and was successfully transmitted from the C. edule at lower temperatures.
Conclusions
This study demonstrates that OsHV-1 μVar has the plasticity to infect the keystone species C. edule and highlights the possible trophic transmission of the virus from cockles to their mobile top predators. This scenario would have important implications, as a greater geographical range expansion of this significant pathogen via migratory bird species may have an impact on other species that reside in bird habitats most of which are special areas of conservation.

Similar content being viewed by others
Background
Coastal and marine ecosystems are routinely used for fisheries and aquaculture purposes [1,2,3] with culture sites often being located on intertidal mud and sandbanks, which have a diverse community of marine invertebrates, algae and vertebrates, including protected wading bird species [4]. Such habitats support a complex interwoven food web with a number of trophic levels. Pathogens and diseases are common in marine habitats [5], often as a result of a complex interplay between the natural host, its environment and the pathogen [6], which can have a significant impact on fisheries and aquaculture due to associated intensive culture [5, 7].
Cerastoderma edule is widely distributed in intertidal ecosystems in North Africa and Europe [8, 9] and is of commercial importance particularly in the British Isles, The Netherlands, Spain and France [8,9,10,11]. Moreover, C. edule plays a significant ecological role as an ecosystem engineer [8, 12, 13], by directly and/or indirectly altering the marine ecosystem. Using its physical structure, C. edule can directly shape its marine environment by forming reefs [13, 14] and as bioturbators it can mobilize sediments and affect sediment stability and accumulation [13, 15,16,17]. Cerastoderma edule functions as a link between primary producers by being a suspension feeder and therefore indirectly change levels of algae, nutrients/metals and sediments [17,18,19,20]. It can also indirectly affect higher trophic levels [20, 21] by acting as an important food source for cohabiting marine species such as crustaceans (the shore crab Carcinus maenas [22] and brown shrimp Crangon crangon [23]), fish species (the European plaice Pleuronectes platessa [24]) and protected wading bird species such as the curlew Numenius arquata and the oystercatcher Haematopus ostralegus [25].
Cockle mortalities have been reported sporadically [26,27,28,29,30] caused by predation, unfavourable environmental circumstances, overfishing, but have also been linked with pathogens and diseases [31]. Cerastoderma edule provides a habitat to a wide range of pathogens and diseases including viruses, bacteria, fungi (Microsporidia), Apicomplexa, Amoeba, Ciliophora, Perkinsozoa, Haplosporidia, Cercozoa, Turbellaria, Digenea, Cestoda, Nematoda, Crustacea and Nemertea [8].
One important global aquaculture bivalve species is the Pacific oyster Crassostrea gigas [32] that cockles often share intertidal beds with. Crassostrea gigas culture has been affected by significant mortalities, associated with ostreid herpesvirus-1 (OsHV-1) and its variants including ostreid herpesvirus-1 microVar (OsHV-1 μVar) since the 1990’s [33,34,35]. Past studies have investigated the possibility of other marine invertebrates being carriers and or reservoirs of OsHV-1 and OsHV-1 μVar, outside the known host C. gigas. A herpes-type infection, the first to be detected in an invertebrate, was observed in the eastern oyster Crassostrea virginica experimentally [36]. In that study, a higher prevalence of the virus occurred in oysters when seawater temperatures were elevated above the normal ambient temperature. A variant of OsHV-1 has been associated with high mortalities of cultured Farrer’s scallops Chlamys farreri in China [37]. López Sanmartín et al. [38] demonstrated that the European flat oyster Ostrea edulis can be infected by OsHV-1 μVar when the virus is administered as an intramuscular injection. More recently OsHV-1 μVar was detected in different marine invertebrates including the Sydney cockle Anadara trapezia in Australia [39], in cultured Mytilus galloprovinciallis in Italy [40], in wild Mytilus spp. consisting of the blue Mytilus edulis, M. galloprovinciallis and hybrids of both parent species, in Ireland [41] and the green shore crab C. maenas [42]. Bookelaar et al. [42] demonstrated that transmission of the virus could occur from C. maenas, previously exposed to the OsHV-1 μVar in the wild, to cohabiting naïve C. gigas within four days. These findings provided some insight on the potential for pathogen species to jump host and more importantly the possibility that this virus can be transmitted through marine food webs and dispersed over greater distances via mobile top predators.
As the cockle plays a significant ecological role, this study investigated the potential of this keystone species as a host, carrier or reservoir of infection for this significant pathogen. The ultimate focus of this study was to shed new light on the potential impacts of anthropogenic introduced pathogens in a wider ecological context.
Methods
The study was conducted in two parts; the first was a field study to screen cockles from areas where oyster culture was taking place for the presence of OsHV-1 μVar and the second part was a laboratory transmission trial to determine if virus could be detected in C. edule from an OsHV-1 μVar endemic Pacific oyster culture site and if transmission could occur to naïve C. gigas.
Field study sites
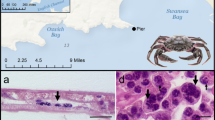
This study took place at the two main Irish oyster culture sites; Dungarvan, Co. Waterford (52.0704°N, 7.5939°W), which covers an area of approximately 25 km2 and Carlingford Lough, Co. Louth (54.0733°N, 6.1994°W), which covers an area of approximately 50 km2 [42] (Fig. 1) [42,43,44]. For both culture sites OsHV-1 μVar has been present since 2009 [45].
In Ireland, oyster culture sites including Dungarvan and Carlingford Lough are known as Special Protection Areas (SPA) under the EU Birds and Habitats Directives as they support over 20,000 waterbirds during the non-breeding season and are of specific conservation interest and importance for bird species [43, 44]. Mobile predators like birds (the oystercatcher Haematopus ostralegus and the common tern Sterna hirundo) are observed in Irish coastal areas year round and are known to prey on C. edule 10–30 mm in shell length, on the upper shore [46].
Environmental data
Environmental data loggers (Star-Oddi) supplied by Bord Iascaigh Mhara (BIM) were used to measure and record continuous water temperature values (every half hour) from the end of May until mid-August 2015 for both sites. Due to a technical issue, data were not recorded from the end of June to the end of July at Dungarvan.
Bivalve sampling
In this study, surfaced (gaping and non-gaping) cockles were randomly sampled at both C. gigas culture sites. Taking into consideration the potential extension range of the virus in the intertidal zone (e.g. by transport of infected particles through the water column), cockles were sampled around the oyster trestles (main culture site) during low tide and approximately 500-m higher up the shore. Thirty cockles were taken at the high shore at both sites and at the oyster trestles in Carlingford Lough (as no surfaced cockles were observed at the trestles in Dungarvan). In addition, to detect baseline levels of virus in the natural host, at every sampling date, 30 C. gigas, originally imported from French hatcheries, were collected at the oyster trestles at both sites.
Cockles were collected biweekly nine times in Dungarvan (April 2015 to August 2015) and eight times in Carlingford Lough (April 2015 to August 2015). As cockles were not present at the trestles in Dungarvan, cockles (250 individuals in total) were only collected at the high shore. At Carlingford Lough, 445 cockles were collected for screening, 218 cockles at the high shore and 227 cockles at the trestles.
Cockles and oysters were processed for histology and molecular analysis on return to the laboratory.
Laboratory transmission trial
Naïve C. gigas (n = 150, average weight of 3.0 g and average length of 31.3 mm), which had never been exposed to OsHV-1 μVar, were obtained from an Irish hatchery in October 2015. Crassostrea edule (n = 93, average weight of 12.6 g and average shell length of 29.9 mm) were randomly collected in October 2015 from Carlingford Lough, where OsHV-1 μVar had been detected in oysters. Prior to the start of the trial, 30 naïve C. gigas and 30 C. edule were screened for OsHV-1 μVar by polymerase chain reaction (PCR), to confirm the oysters were uninfected and to determine if the virus could be detected in C. edule.
Before placing in tanks, the exterior of the shells of C. edule were washed several times in ddH2O to remove any pathogens that may have been incidentally attached to their external shell. Four 10-l tanks were filled with 8-l UV filtered natural seawater and animals were held at 14 ℃ in a constant temperature (CT) room with a salinity of 35. The experimental set up consisted of a control tank containing 30 naïve oysters and three replicate experimental tanks, which each contained 30 naïve C. gigas and 21 C. edule. The tanks were checked several times daily for mortality and any gaping and/or dead individuals were removed to screen for OsHV-1 μVar by PCR. In addition, oysters (n = 3) were sampled on Day 2 (48 hours), Day 4 (96 hours) and Day 7 (168 hours), to screen the naïve oysters for the virus to determine if any replication of the virus was taking place by assessing viral loads. All individuals, C. gigas and C. edule, still alive at the end of the experiment were removed and screened for OsHV-1 μVar. Since most cockles died within the first few days the trial was terminated at 14 days.
Cockle sample processing
Whole wet weight (g) and shell length (mm) of cockles from both field and laboratory trials were recorded using a balance scales and Vernier calipers. In addition, shell rings were counted. Shell lengths were divided into five different length classes: Class 1: 0–8 mm; Class 2: 8.01–16.00 mm; Class 3: 16.01–24.00 mm; Class 4: 24.01–32.00 mm; and Class 5: 32.01–40.00 mm. Weights were categorized as four weight classes: Class 1: 0–7 g; Class 2: 7.01–14.00 g; Class 3: 14.01–21.00 g; and Class 4: 21.01–28.00 g. Growth rings on the shell were divided into seven different shell ring classes: Class 1: 0 rings; Class 2: 1 ring; Class 3: 2 rings; Class 4: 3 rings; Class 5: 4 rings; Class 6: 5 rings; and Class 7 > 5 rings. Correlation tests were performed between weight and age and between shell length and age to clarify if growth rings could be associated with life span.
DNA extraction
Gill and internal tissues (approx. 5 mm2) made up of connective, digestive and reproductive tissues of C. edule and gill tissue of C. gigas, from both field and laboratory trials, were stored in 95% ethanol for DNA isolation to detect OsHV-1 μVar. Before DNA extraction took place, tissues were washed in double deionized water (ddH2O) and blot dried using tissue paper. DNA extraction was performed using the Chelex-100 methodology [47]. Tissue samples were placed in a 10% chelex solution (100 µl volume) (Sigma Aldrich) and following the samples were placed in a Hybaid thermal cycler for 1 h and 10 min heated at 99 ℃ to facilitate cell lysis [47]. A subsample (n = 30) of the cockle DNA was quantified by a spectrophotometer (NanoDrop 1000 spectrophotometer) to confirm DNA quantity and quality to avoid the diagnosis of false negatives due to low DNA quantity and/or DNA of poor quality. The ratio of absorbance at 260 nm and 280 nm was used to assess the purity of DNA with a ratio of ~ 1.8 considered “pure” for DNA.
Polymerase chain reaction (PCR)
Standard PCR to detect OsHV-1 μVar was performed using the OHVA/OHVB primers [48]. Positive controls (duplicate) consisting of OsHV-1 μVar infected oyster tissue and negative controls (duplicate) of double distilled water (ddH2O) were used in duplicate for each PCR. Agarose gel electrophoresis was carried out using a 2% agarose gel stained with ethidium bromide (10 mg/l stock) and was run with an electrical charge of 110V for 45–60 min. Expected size of amplified PCR products for OsHV-1 μVar was 385 bp [48].
Quantitative polymerase chain reaction (qPCR)
Quantitative PCR (qPCR) was carried out to determine the viral load of a subsample of cockles from the field trial (n = 76), including gill (n = 33) and internal tissue (n = 43) of cockles from both sites, deemed positive for OsHV-1 μVar by standard PCR. In addition, qPCR was performed on C. edule and C. gigas from the laboratory transmission trial, which were deemed positive for OsHV-1 μVar by standard PCR. qPCR conditions, master mix and thermocycling conditions were carried out using the HVDP-F and HVDP-R primers [49]. Samples were tested in duplicate and samples with a mean CT value of below 37 were indicative of a positive result for the virus.
In situ hybridization (ISH)
In situ hybridization assays (ISH) were carried out on cockles from the field survey that were negative (n = 3 for both sites) and positive (n = 3 for both sites) for OsHV-1 μVar when screened by PCR and qPCR. In addition, ISH was performed on infected (n = 3) and uninfected (n = 3) C. gigas from the laboratory trial. An ISH protocol that has been used to screen for Bonamia ostreae was carried out using a digoxigenin (DIG)-labelled probe specific to OsHV-1 μVar [50].
Sanger sequencing
Sequencing of genomic DNA from positive PCR products amplified from C. edule (n = 5) from the field screening (Carlingford (n = 3) and Dungarvan (n = 2)) took place. PCR products in triplicate from each individual were pooled together to increase DNA concentration prior to being sent for direct sequencing to Eurofins MWG. DNA sequences that were generated by Eurofins MWG were matched against the National Center for Biotechnology Information (NCBI) nucleotide database with BLASTn to identify and confirm that the sequences were specifically OsHV-1 μVar strains.
Statistical analyses
Statistical analyses were performed in statistical model program R studio [51]. Normality was tested using the Shapiro-Wilks normality test. Environmental data between sites were analysed by Studentʼs t-test. Morphometrics (length and weight) between sites and shore heights were analysed by Mann-Whitney U-test. Spearmanʼs rank order correlation was used to determine if there was an association between shell rings and weight and between shell rings and length. Pearsonʼs Chi-square tests were used to compare length and weight classes and number of shell rings between sites and within sites at the two shore heights. Pearsonʼs Chi-square tests were also used to test for differences of prevalence of OsHV-1 μVar for gill and internal tissue for cockle within sites at the two shore heights. For all analyses, a critical value of 0.05 was used to confirm significant results. Data are presented as the mean ± standard error.
Results
Baseline levels of OsHV-1 μVar at Pacific oyster culture sites
OsHV-1 μVar was present in oysters at the sample sites. Overall prevalence of OsHV-1 μVar by PCR in C. gigas at Dungarvan (n = 270) and Carlingford Lough (n = 240) for the duration of the field trial was low (< 10%) with a mean prevalence of 6.0% and a range of 0–27% per month at Dungarvan and a mean prevalence of 6.0% with a range of 0–23% per month at Carlingford Lough (Fig. 2). The overall water temperature during summer 2015 remained low (< 16 ℃).
Virus detection in Cerastoderma edule in the field
OsHV-1 μVar was detected in the cockles at both sample sites, and in both gill and internal tissues in the PCR screening. A subsample of cockle DNA (n = 30) was screened for DNA quantity and quality using a NanoDrop spectrophotometer. Regarding quantity all samples showed a sufficient amount of DNA higher than 40 μg/l and 80% of the samples had DNA higher than 100 μg/l. 260/280 ratio indicated values of 1.5–2.1. 260/230 ratios represented values ranging between 1.5 and 2.0, representing low contamination levels.
The overall prevalence of OsHV-1 μVar in C. edule for Dungarvan was 14.4% (14.4% at high shore) and in Carlingford Lough was 13.6% (12.8% at high shore and 14.3% at trestles). More individuals demonstrated the presence of the virus in the internal tissues (prevalence of 10.2%; 71/695) exclusively, followed by individuals with infection in gill only and individuals with infection in both tissues (Table 1). This pattern however was only significantly different at the high shore at Carlingford Lough (χ2 = 12.43, df = 2, P < 0.01) and not at the high shore at Dungarvan (χ2 = 2.29, df = 2, P = 0.32) and at the trestles at Carlingford Lough (χ2 = 1, df = 2, P = 0.61) (Table 1).
Significant differences in prevalence between months were observed for both Dungarvan (χ2 = 25.15, df = 4, P < 0.01) and Carlingford Lough (χ2 = 17.95, df = 4, P < 0.01) with highest prevalence in May and lowest prevalence in April at both sites (Fig. 2). No significant trends were observed for herpesvirus prevalence and the size and age of the cockles (indicated by growth rings), with the virus being detected in all length and weight classes and all shell ring classes (Fig. 3).
Viral load in cockle tissues was generally low with the majority of cockles sampled having less than 102 and a small number having 103 and 104 viral copies (Fig. 4). Confirmation of the presence of OsHV-1μVar was carried out using ISH; a positive signal was observed for a small number of C. edule (83.3%; 5/6) screened, these had been collected in the field, for the laboratory trial (Fig. 5).
After sequencing of the PCR products of C. edule (n = 5), BLASTn analysis showed a match with OsHV-1 μVar for the sequence of the PCR-amplified products of C. edule with an average of 92.6% (88–95%) query coverage and 99% maximum identity to OsHV-1 μVar (GenBank: KU861511.1).
Laboratory cohabitation transmission trial
Transmission was effected from cockles infected with OsHV-1μVar to naïve uninfected oysters in the laboratory. Prior to the trial commencing (collected in October 2015), 30 individuals of each species (cockles and oysters) were screened. Naïve C. gigas were uninfected with OsHV-1 μVar as expected. The initial sample of C. edule had a prevalence of 6.7% (n = 2/30) with a mean viral load of 1.1 × 101 copies/μl of genomic DNA (range: < 10–3.0 × 101 viral copies/μl of genomic DNA). Overall, the prevalence of infection in the C. edule held with the C. gigas in a single experimental tank was 3.2% (n = 2/63) with an average viral DNA load of 1.2 × 101 copies/μl (range: < 10–1.2 × 104 viral copies/μl of genomic DNA). OsHV-1 μVar was detected in a single dead oyster on Days 5, 8, 11 and 13. C. gigas held with C. edule in that single experimental tank had an overall OsHV-1 μVar prevalence of 4.4% (n = 4/90) with an average viral load of 3.0 × 103 DNA viral copies/ μl of genomic DNA (range: < 10–1.2 × 104 viral copies/μl of genomic DNA). No OsHV-1 μVar was observed in the C. edule or C. gigas in the two additional experimental tanks/replicates.
During the laboratory trial, cumulative mortality of up to 100% was observed in C. edule in all three replicate experimental tanks within the first 4 days of the trial (n = 63). Crassostrea gigas showed low mortality up to Day 7 in all three experimental tanks (< 10%), after Day 7 mortality increased up to 66.7–100% (with a mean of 85.7% for all three experimental tanks) at the end of the trial (Fig. 6). All C. gigas in the control tank were still alive at the end of the trial.
Discussion
In this study, the potential for an intertidal ecosystem engineer C. edule to become infected with and to act as an alternate host of OsHV-1 μVar was determined, through field screening, laboratory transmission trials and using a variety of diagnostic methods. The field survey demonstrated the ability of a virus to jump host to an alternate infaunal bivalve with potential further transmission through an ecosystem and potentially greater geographic dispersal via trophic food webs, as was demonstrated by Bookelaar et al. [42]. In that study, OsHV-1 μVar was detected in C. maenas, a predator/scavenger of C. gigas and C. edule [52], and transmission of the virus occurred between infected crabs and naïve C. gigas. This suggests that OsHV-1 μVar is exhibiting plasticity, by spreading to other bivalves. Detection of OsHV-1 μVar did not show a significant correlation with cockle size, weight and age (number of shell rings), which indicated that all cockle cohorts were likely to be infected with the virus. In addition, the laboratory trial indicated that cockles can transmit viable OsHV-1 μVar back to oysters within five days post-exposure and at a low temperature of 14 ℃, below the temperature (16 ℃) considered necessary for activation and replication of the virus. The mortality of all C. edule by the fourth day of the trial in all three experimental tanks (one with infected C. edule and two with uninfected C. edule) was not exclusively due to OsHV-1 μVar but possibly due to the artificial holding conditions or another health condition. Cerastoderma edule typically do not perform well (i.e. survive) in artificial holding conditions in particular stand-alone tanks (our unpublished data). Additionally, a substrate was not provided, which may have acted as a stressor by preventing their normal burrowing behaviour. This was a deliberate step to encourage the C. edule, if infected, to shed the virus into the tank system holding the C. gigas.
Herpesvirus infection was detected in the oysters being cultured at the sites in this study, but at a lower level compared to previous years in Ireland [35, 45, 53]. In this study, oysters selectively bred for resistance to OsHV-1 μVar were used (oyster farmers, personal communications), which might be the main explanation for the lower prevalence in oysters. Moreover, temperatures were lower compared with previous years and rarely passed the temperature level (16 ℃) considered necessary for OsHV-1 μVar to activate, but the subsequent laboratory trial demonstrated the transmission could occur at 14 ℃ from cockles to oysters. Survival of aquatic viruses such as Abalone herpes virus outside their natural hosts, has been mostly observed at lower temperatures [54]. In addition, a previous study investigating prevalence of OsHV-1 in Ostrea edulis observed a lower optimal temperature of 6–12 ℃ for O. edulis, compared to the threshold temperature of 16 ℃ in C. gigas [55]. In addition, a 14-fold OsHV-1 μVar replication was observed in mussels Mytilus spp. at 13 ℃ under experimental conditions [41]. The findings of this study would suggest that OsHV-1 μVar may be opportunistic, adapting to new circumstances, such as fewer available susceptible oysters due to the use of selectively bred oyster stocks more resistant to the virus, resulting in a “species jump” to another cohabiting bivalve species, and being transmissible at lower seawater temperatures.
A high percentage of the cockles showed lower viral copies than the viral copy number of 104 mg−1 associated with oyster mortalities [56]. This phenomenon was also found in a recent Australian study where low viral copies of OsHV-1 μVar were detected in other marine invertebrates; mussels, whelks and barnacles and it was suggested that these species could function as reservoirs and/or support transmission of OsHV-1 μVar in the environment [39]. However, that study did not investigate if those infected invertebrates could transmit the virus to oysters. Of significance in this study, a few individuals did have a viral copy number of 104 mg−1 cockle tissue, which would indicate replication of the virus in the cockles.
Conclusions
As a possible consequence of this ‘species jump’, and finding the virus in all surfaced cockle sizes/ages, it is likely infected cockles could be consumed by mobile top predators more readily, like bird and fish species, facilitating the transmission and greater geographical dispersal of the virus and potentially transmitting the virus to new carriers, reservoirs or possibly new hosts. The potential of OsHV-1 μVar to extend its geographical range and be introduced to uninfected marine habitats is possible with the migration of such mobile predators. Certainly, previous studies indicated that birds contribute to geographical jumps of emerging infectious diseases and have been proven carriers of Lyme disease and influenza A [57] and white spot disease in penaeid shrimps Penaeus monodon [58]. Changes in environmental conditions, in particular warming seas, are predicted to create a more conducive environment for viral replication. Such changes might result in significant cockle mortalities and as a keystone species any negative impacts through increased mortality or reduced physiological ability are likely to have wider ecosystem and commercial impacts.
Availability of data and materials
Data supporting the conclusions of this article are included within the article. The datasets generated and analysed during the present study are available from the corresponding author upon reasonable request.
References
Hattam C, Atkins JP, Beaumont N, Börger T, Böhnke-Henrichs A, Burdon D, et al. Marine ecosystem services: linking indicators to their classification. Ecol Indic. 2015;49:61–75.
FAO. Global capture production database updated to 2015 - summary information; Rome: Food and Agriculture Organization; 2017. http://www.fao.org/3/a-br186e.pdf.
FAO. An overview of recently published global aquaculture statistics updated to 2015—summary information. Rome: Food and Agriculture Organization; 2017. http://www.fao.org/3/a-bs235e.pdf. Accessed 28 Aug 2020.
European Commission DG Environment. Interpretation Manual of European Union Habitats—EUR28. 2013. http://ec.europa.eu/environment/nature/legislation/habitatsdirective/docs/Int_Manual_EU28.pdf. Accessed 28 Aug 2020.
Lafferty KD, Harvell CD, Conrad JM, Friedman CS, Kent ML, Kuris AM, et al. Infectious diseases affect marine fisheries and aquaculture economics. Ann Rev Mar Sci. 2015;7:471–96.
Engering A, Hogerwerf L, Slingenbergh J. Pathogen-host-environment interplay and disease emergence. Emerg Microbes Infect. 2013;2:5.
Willman R, Kelleher K, Arnason R, Franz N. The sunken billions: the economic justification for fisheries reform. Washington, DC: World Bank and FAO; 2009.
Longshaw M, Malham SK. A review of the infectious agents, parasites, pathogens and commensals of European cockles (Cerastoderma edule and C. glaucum). J Mar Biol Assoc UK. 2013;93:227–47.
Dare PJ, Bell MC, Walker P, Bannister RCA. Historical and current status of cockle and mussel stocks in the Wash. Lowestoft: CEFAS; 2004.
Carrasco N, Rojas M, Aceituno P, Andree KB, Lacuesta B, Furones MD. Perkinsus chesapeaki observed in a new host, the European common edible cockle Cerastoderma edule, in the Spanish Mediterranean coast. J Invertebr Pathol. 2014;117:56–60.
11. FAO. Global aquaculture production 1950–2014 database. Rome: Food and Agriculture Organization; 2017. http://www.fao.org/fishery/statistics/en. Accessed 28 Aug 2020.
Morgan E, O’Riordan RM, Kelly TC, Culloty SC. Influence of disseminated neoplasia, trematode infections and gametogenesis on surfacing and mortality in the cockle Cerastoderma edule. Dis Aquat Org. 2013;98:73–84.
Donadi S, van der Heide T, Piersma T, van der Zee EM, Weerman EJ, van de Koppel J, et al. Multi-scale habitat modification by coexisting ecosystem engineers drives spatial separation of macrobenthic functional groups. Oikos. 2015;124:1502–10.
Donadi S, van der Heide T, van der Zee EM, Eklöf JS, van de Koppel J, Weerman EJ, et al. Cross-habitat interactions among bivalve species control community structure on intertidal flats. Ecology. 2013;94:489–98.
Montserrat F, Provoost C, Van Colen P, Milla M, Ponti M, Van den Meersche K, et al. Sediment segregation by biodiffusing bivalves. Estuar Coast Shelf Sci. 2009;83:379–91.
Donadi S, Westra J, Weerman EJ, van der Heide T, van der Zee EM, van de Koppel J, et al. Non-trophic interactions control benthic producers on intertidal flats. Ecosystems. 2013;16:1325–35.
Eriksson BK, Westra J, van Gerwen I, Weerman E, van der Zee E, van der Heide T, et al. Facilitation by ecosystem engineers enhances nutrient effects in an intertidal system. Ecosphere. 2017;8:e02051.
Eriksson BK, van der Heide T, van de Koppel J, Piersma T, van der Veer HW, Olff H. Major changes in the ecology of the Wadden Sea: human impacts, ecosystem engineering and sediment dynamics. Ecosystems. 2010;13:752–64.
Jones CG, Lawton JH, Shachak M. Organisms as ecosystem engineers. Oikos. 1994;69:373–86.
Cesar CP, Frid CLJ. Effects of experimental small-scale cockle (Cerastoderma edule L.) fishing on ecosystem function. Mar Ecol. 2009;30:123–37.
Lotze HK. Radical changes in the Wadden Sea fauna and flora over the last 2000 years. Helgol Mar Res. 2005;59:71–83.
Beukema JJ. The abundance of shore crabs Carcinus maenas (L.) on a tidal flat in the Wadden Sea after cold and mild winters. J Exp Mar Biol Ecol. 1991;153:97–113.
Beukema JJ. Dynamics of juvenile shrimp Crangon crangon in a tidal-flat nursery of the Wadden Sea after mild and cold winters. Mar Ecol Prog Ser. 1992;83:157–65.
Pihl L. Food selection and consumption of mobile epibenthic fauna in shallow marine areas. Mar Ecol Prog Ser. 1985;2:169–79.
Goss-Custard JD, Jones RE. The diets of redshank and curlew. Bird Study. 1976;23:233–43.
Rybarczyk H, Elkaim B, Wilson JG, Loquet N. Eutrophication in the Baie de Somme: consequences and impacts on the benthic population. Oceanologica Acta. 1996;19:131–40.
Masski H, Guillou J. The role of biotic interactions in juvenile mortality of the cockle (Cerastoderma edule L.): field observations and experiment. J Shellfish Res. 1999;18:575–8.
Atkinson PW, Clark NA, Clark JA, Bell MC, Dare PJ, Ireland PL. Changes in commercially fished shellfish stocks and shorebird populations in the Wash, England. Biol Conserv. 2003;114:127–41.
Beukema JJ, Dekker R, Philippart CJM. Long-term variability in bivalve recruitment, mortality, and growth and their contribution to fluctuations in food stocks of shellfish-eating birds. Mar Ecol Prog Ser. 2010;414:117–30.
Ducrotoy JP, Rybarczyk H, Souprayen J, Bachelet G, Beukema JJ, Desprez M, et al. A comparison of the population dynamics of the cockle (Cerastoderma edule L.) in north-western Europe. Estuar Coast Shelf Sci. 1991;19:173–84.
Thieltges DW. Parasite induced summer mortality in the cockle Cerastoderma edule by the trematode Gymnophallus choledochus. Hydrobiologia. 2006;559:455–61.
FAO. Fisheries and Aquaculture Fact Sheets. Rome: food and agriculture organization; 2017. http://www.fao.org/fishery/species/3514/en. Accessed 28 Aug 2020.
Burge CA, Judah LR, Conquest LL, Griffin FJ, Cheney DP, Suhrbier A, et al. Summer seed mortality of the Pacific oyster, Crassostrea gigas Thunberg grown in Tomales Bay, California, USA: the influence of oyster stock, planting time, pathogens, and environmental stressors. J Shellfish Res. 2007;26:163–72.
Lynch SA, O’Reilly A, Cotter E, Carlsson J, Culloty SC. A previously undescribed ostreid herpes virus (OsHV-1) genotype detected in the Pacific oyster, Crassostrea gigas, in Ireland. Parasitology. 2012;139:1526–32.
Prado-Alvarez M, Darmody G, Hutton S, O’Reilly A, Lynch SA, Culloty SC. Occurrence of OsHV-1 in Crassostrea gigas cultured in Ireland during an exceptionally warm summer. Selection of less susceptible oysters. Front Physiol. 2016;7:492.
Farley CA, Banfield WG, Kasnic G Jr, Foster WS. Oyster herpes-type virus. Science. 1972;178:759–60.
Ren W, Chen H, Renault T, Cai Y, Bai C, Wang C, Huang J. Complete genome sequence of acute viral necrosis virus associated with massive mortality outbreaks in the Chinese scallop, Chlamys farreri. Virol J. 2013;10:110.
López Sanmartín M, Power DM, de la Herrán R, Navas JI, Batista FM. Experimental infection of European flat oyster Ostrea edulis with ostreid herpesvirus 1 microvar (OsHV-1 var): mortality, viral load and detection of viral transcripts by in situ hybridization. Virus Res. 2016;217:55–62.
Evans O, Paul-Pont I, Whittington RJ. Detection of ostreid herpesvirus 1 microvariant DNA in aquatic invertebrate species, sediment and other samples collected from the Georges River estuary, New South Wales, Australia. Dis Aquat Org. 2017;122:247–55.
Domeneghetti S, Varotto L, Civettini M, Rosani U, Stauder M, Pretto T, et al. Mortality occurrence and pathogen detection in Crassostrea gigas and Mytilus galloprovincialis close-growing in shallow waters (Goro Lagoon, Italy). Fish Shellfish Immunol. 2014;41:37–44.
OʼReilly AJ, Laide C, Maloy A, Hutton S, Bookelaar B, OʼSullivan K, et al. The role of the mussel Mytilus spp. in the transmission of ostreid herpesvirus-1 microVar. Parasitology. 2017;145:1095–104.
Bookelaar B, O’Reilly A, Lynch SA, Culloty SC. Role of the intertidal predatory shore crab Carcinus maenas in transmission dynamics of ostreid herpesvirus-1 microvariant. Dis Aquat Org. 2018;130:221–33.
NPWS (National Parks and Wildlife Service). 2011. Dungarvan Harbour Special Protection Area (Site Code 4032). Conservation Objectives Supporting Document. https://www.npws.ie/sites/default/files/publications/pdf/004032_Dungarvan%20Harbour%20SPA%20Supporting%20Doc_V1.pdf. Accessed 28 Aug 2020.
NPWS (National Parks and Wildlife Service). 2013. Carlingford Lough Special Protection Area (4078). Conservation objectives supporting document. https://www.npws.ie/sites/default/files/publications/pdf/004078_Carlingford%20Lough%20SPA%20Supporting%20Doc_V1.pdf. Accessed 28 Aug 2020.
Morrissey T, McCleary S, Collins E, Henshilwood K, Cheslett D. An investigation of ostreid herpes virus microvariants found in Crassostrea gigas oyster producing bays in Ireland. Aquaculture. 2015;442:86–92.
Reise K. Tidal flat ecology - an experimental approach to species interactions. Berlin: Springer-Verlag; 1985.
Walsh PS, Metzger DA, Higuchi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 1991;10:506–13.
Lynch SA, Dillane E, Carlsson J, Culloty SC. Development and assessment of a sensitive and cost-effective polymerase chain reaction to detect ostreid herpesvirus 1 and variants. J Shellfish Res. 2013;32:1–8.
Webb SC, Fidler A, Renault T. Primers for PCR-based detection of ostreid herpes virus-1 (OsHV-1): application in a survey of New Zealand molluscs. Aquaculture. 2007;272:126–39.
Lynch SA, Abollo E, Ramilo A, Cao A, Culloty SC, Villalba A. Observations raise the question if the Pacific oyster Crassostrea gigas can act as either a carrier or a reservoir for Bonamia ostreae or Bonamia exitiosa. Parasitology. 2010;137:1515–26.
R Core Development Team. R: a language and environment for statistical computing. Vienna: R Foundation for Statistical computing. https://www.R-project.org/.
Mascaró M, Seed R. Choice of prey size and species in Carcinus maenas (L.) feeding on four bivalves of contrasting shell morphology. Hydrobiologia. 2001;449:159–70.
Clegg TA, Morrissey T, Geoghegan F, Martin SW, Lyons K, Ashe S, More SJ. Risk factors associated with increased mortality of farmed Pacific oysters in Ireland during 2011. Prev Vet Med. 2014;113:257–67.
Corbeil S, McColl KA, Williams LM, Mohammad I, Hyatt AD, Crameri SG, et al. Abalone viral ganglioneuritis: establishment and use of an experimental immersion challenge system for the study of abalone herpes virus infections in Australian abalone. Virus Res. 2012;165:207–13.
Feist S. Report of the Annual Meeting of the National Reference Laboratories for Mollusc Diseases, 18–19 March 2008, Nantes. France. https://www.eurl-mollusc.eu/content/download/36313/493331/file/. Accessed 28 Aug 2020.
Oden E, Martenot C, Travaille E, Malas JP, Houssin M. Quantification of ostreid herpesvirus 1 (OsHV-1) in Crassostrea gigas by real-time PCR: determination of a viral load threshold to prevent summer mortalities. Aquaculture. 2011;317:27–31.
Reed KD, Meece JK, Henkel JS, Shukla SK. Birds, migration and emerging zoonoses: West Nile virus, Lyme disease, influenza A and enteropathogens. J Clin Med Res. 2003;1:5–12.
Altizer S, Bartel R, Han BA. Animal migration and infectious disease risk. Science. 2011;331:296–302.
Acknowledgements
This study was undertaken in the Reposus Project 14/SF/820 as part of the Food Institutional Research Measure (FIRM) funded by the Irish Department of Agriculture, Food and the Marine, and the Cockles Project funded by the INTERREG Atlantic Area European Programme grant number EAPA_458/2016. We would like to thank the Pacific oyster growers for allowing us access to their culture sites for the cockle sampling and for providing us with the oyster samples.
Funding
This study was funded in part by the Food Institutional Research Measure (FIRM) Programme, Irish Department of Agriculture, Food and the Marine, and by the INTERREG Atlantic Area European Programme grant number EAPA_458/2016.
Author information
Authors and Affiliations
Contributions
BB carried out the field work, laboratory trial, sample analysis and the writing of the manuscript. SL assisted with the experimental design, field work, sample analysis and the writing of the manuscript. SCC provided the experimental design and the writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study complies with institutional, national and European animal ethics guidelines and policy (EU Code 2017) and was approved by the institutional ethics committee at University College Cork.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Bookelaar, B., Lynch, S.A. & Culloty, S.C. Host plasticity supports spread of an aquaculture introduced virus to an ecosystem engineer. Parasites Vectors 13, 498 (2020). https://doi.org/10.1186/s13071-020-04373-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-020-04373-y