Abstract
Background
Little is known about the prevalence and zoonotic potential of Cryptosporidium spp. and Giardia duodenalis in deer in China. In this study, 662 fecal samples were collected from 11 farms in Henan and Jilin Provinces between July 2013 and August 2014, and were screened for the presence of Cryptosporidium and G. duodenalis with genotyping and subtyping methods.
Results
Cryptosporidium spp. and G. duodenalis were detected in 6.80% (45/662) and 1.21% (5/662) of samples, respectively. Six Cryptosporidium species/genotypes were identified based on the small subunit ribosomal ribonucleic acid (SSU rRNA) gene: C. parvum (n = 11); C. andersoni (n = 5); C. ubiquitum (n = 3); C. muris (n = 1); C. suis-like (n = 1); and Cryptosporidium deer genotype (n = 24). When five of the 11 C. parvum isolates were subtyped by sequencing the 60 kDa glycoprotein (gp60) gene, zoonotic subtypes IIaA15G2R2 (n = 4) and IIdA19G1 (n = 1) were found. According to a subtype analysis, three C. ubiquitum isolates belonged to XIIa subtype 2. In contrast, only assemblage E was detected in the five Giardia-positive samples with small subunit ribosomal ribonucleic acid (SSU rRNA) gene sequencing.
Conclusions
To our knowledge, this is the first study to report C. andersoni, as well as C. parvum zoonotic subtypes IIaA15G2R2 and IIdA19G1 in cervids. These data, though limited, suggest that cervids may be a source of zoonotic Cryptosporidium and Giardia. Cervids in the present study are likely to be of low zoonotic potential to humans, and more molecular epidemiological studies are required to clarify the prevalence and public health significance of Cryptosporidium and G. duodenalis in cervids throughout China.
Similar content being viewed by others
Background
Cryptosporidium and Giardia are two common protozoan parasites responsible for diarrhea in a broad range of vertebrate hosts, including humans, and domestic and wild animals worldwide. Transmission of both pathogens is by the fecal-oral route with both zoonotic and anthroponotic transmission cycles [1, 2]. The host plays an important role in the clinical impact of Cryptosporidium and G. duodenalis infections and the expression of disease. Drug treatments for these infections are inadequate, and do not provide a reliable strategy for their control [3].
Molecular epidemiological research into deer Cryptosporidium has been undertaken in red deer (Cervus elaphus), roe deer (Capreolus capreolus), swamp deer (Rucervus duvaucelii), sika deer (Cervus nippon), fallow deer (Dama dama), sambar deer (Rusa unicolor), caribou (Rangifer tarandus), white-tailed deer (Odoileus virginianus) and black-tailed deer (Odocoileus hemionus), in Europe (Spain, the Czech Republic and the UK), Asia (Nepal, Japan and China), Canada, the USA and Australia [4,5,6,7,8,9,10,11,12,13,14,15,16,17,18]. So far, 11 Cryptosporidium species/genotypes have been identified in cervids: C. parvum, C. hominis, C. bovis, C. ryanae, C. ubiquitum, C. muris, Cryptosporidium deer genotype, Cryptosporidium suis-like genotype, Cryptosporidium muskrat II genotype, C. hominis-like genotype and Cryptosporidium caribou genotype. Giardia duodenalis is considered a species complex that infects humans and many other mammals. Eight genetic groups or assemblages (A-H) have been identified based on a variety of genetic loci: assemblages A and B occur in humans and many other mammals; assemblages C and D in dogs; assemblage E in artiodactyls; assemblage F in cats; assemblage G in rodents; and assemblage H in seals [1]. However, only a few studies have reported the molecular characterization of G. duodenalis in cervids, with assemblages A, B, D and E identified [6, 13, 16, 19,20,21,22,23,24,25,26,27,28,29].
As an important center of mammalian evolution and dispersal, China possesses an abundance of deer species [30]. Deer and their products are of high economic value and deer farming has become an important component of China’s animal breeding industry. Sika deer, red deer, sambar deer, white-lipped deer, reindeer, Eld’s deer and Père David’s deer are the major species farmed in China. There are approximately 550,000 domesticated sika deer, most of which are distributed in northwestern China [31]. Velvet antlers, important in traditional Chinese medicine, are one of the main products derived from sika deer. Père David’s deer is an endemic species in China, but has become extinct in the wild [32].
Some epidemiological surveys exist concerning Cryptosporidium and Giardia in cervids around the world, but little is known about the prevalence and molecular characteristics of Cryptosporidium and Giardia in cervids in China. Only one study was conducted in Zhengzhou, where 124 fecal specimens were examined and two C. ubiquitum isolates were identified in sika deer [14]. In the present study, deer-derived Cryptosporidium and Giardia isolates were genetically characterized to better understand the distribution and zoonotic potential of the two pathogens in cervids in Henan and Jilin provinces.
Methods
Samples
In total, 662 samples were collected between July 2013 and August 2014 from 11 farms in Henan and Jilin provinces, from 16 red deer (Cervus elaphus), 47 Père David’s deer (Elaphurus davidianus) and 599 sika deer (Cervus nippon) (Table 1). Red deer and sika deer were in shed-feeding and housed in separate breeding houses according to different deer species and age groups. These 615 animals had a wide age distribution, ranging from 1 month to 15 years. However, the Père David’s deer from one forest farm were so agile, solitary and secretive, that it was difficult to determine their precise ages. Approximately 50 g of fresh feces was collected from each deer immediately after its defecation onto the ground, using a sterile disposal latex glove, and was then placed individually into a disposable plastic bag. No obvious clinical signs were observed in these deer, except for one case of diarrhea. The specimens were transported to the laboratory in an insulated container containing cold packs. Upon arrival, a portion of each specimen was examined by microscopy to detect Cryptosporidium oocysts and Giardia cysts using Sheather’s sugar flotation technique and Lugol’s iodine stain method, respectively. Wet smears were examined using a bright-field microscope with 100× and 400× magnification. All of the fecal specimens were stored in 2.5% potassium dichromate solution at 4 °C prior to DNA extraction.
DNA extraction
The fecal specimens were washed three times in distilled water and centrifuged at 3000× g for 10 min to remove the potassium dichromate. DNA was extracted from 200 mg of each fecal specimen using the E.Z.N.A. Stool DNA Kit (Omega Biotek Inc., Norcross, GA, USA), according to the manufacturer’s instructions. The extracted DNA was stored at -20 °C.
Genotyping and subtyping
Cryptosporidium species were identified by nested PCR amplification and sequencing of an ~830 bp fragment of the small subunit ribosomal ribonucleic acid (SSU rRNA) gene, as described previously [2]. Cryptosporidium parvum and C. ubiquitum were subtyped by a sequence analysis of the 60 kDa glycoprotein (gp60) gene [33, 34]. The assemblages of G. duodenalis were determined by sequencing the small subunit ribosomal ribonucleic acid (SSU rRNA), β-giardin (bg), glutamate dehydrogenase (gdh), and triosephosphate isomerase (tpi) genes [35, 36]. Replicate analyses were done at each locus using both positive and negative controls.
Sequence analysis
All PCR amplicons were sequenced on an ABI Prism™ 3730 XL DNA Analyzer (Applied Biosystems, Foster, CA, USA), using the BigDye Terminator v.3.1 Cycle Sequencing Kit (Applied Biosystems). The sequencing accuracy was confirmed by two-directional sequencing. The sequences were identified by their alignment with reference sequences downloaded from GenBank using the MEGA 6.0 software [37]. Representative nucleotide sequences generated in this study have been deposited in the GenBank database under the accession numbers KX259127-KX259145 and MG921620-MG921622.
Statistical analysis
The statistical analysis was performed with the SPSS 22.0 software. Chi-square test and 95% confidence intervals (CIs) were used to compare the Cryptosporidium prevalence rates among different locations and age groups, and differences were considered significant at P < 0.05.
Results
Prevalence
Microscopic analysis of 662 cervine fecal samples showed an identical presence of Cryptosporidium oocysts and Giardia cysts to PCR assay. The overall prevalence of Cryptosporidium spp. in cervids was 6.8% (45/662, 95% CI: 4.9–8.7%). All 11 farms were positive for Cryptosporidium, with prevalences ranging between 4.9–11.5% (Table 1). No difference was observed in the Cryptosporidium prevalences in Jilin (6.91%, 32/463, 95% CI: 4.6–9.2%) and Henan (6.53%, 13/199, 95% CI: 3.1–10.0%) (χ2 = 0.032, df = 1, P > 0.05). Only five Giardia-positive samples were detected (0.76%, 5/662, 95% CI: 0.1–1.4%), from two farms in Henan Province (Table 1). Cryptosporidium was found in red deer, Père David’s deer, and sika deer, but Giardia was only detected in sika deer.
Cryptosporidium species/genotypes
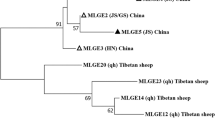
Forty-five Cryptosporidium-positive samples were genotyped by a sequence analysis of the SSU rRNA gene, and six Cryptosporidium species/genotypes were identified: Cryptosporidium deer genotype (n = 24, GenBank: KX259127-KX259129), C. parvum (n = 11, GenBank: KX259136-KX259140), C. andersoni (n = 5, GenBank: KX259130-KX259131), C. ubiquitum (n = 3, GenBank: KX259133-KX259134), C. muris (n = 1, GenBank: KX259132) and C. suis-like (n = 1, GenBank: KX259135). Phylogenetic relationship analysis confirmed the identity of Cryptosporidium species (Fig. 1). The Cryptosporidium deer genotype was the predominant genotype (χ2 = 7.901, df = 1, P < 0.001), and had an identical sequence to that isolated from white-tailed deer (KR260681) in the Czech Republic [10].
When the 11 C. parvum isolates sequences were compared with each other, five variants were detected. Variant 1 was identical to sequence AB513881 derived from calves in Egypt [38], while variants 2–5 had one or two nucleotide substitutions at six sites in the SSU rRNA sequence; these variants have only been found in this study. Two variants of C. andersoni were identified, and variant 1 and variant 2 were identical to sequences KF826313 and KF826314, respectively, isolated from Chinese outpatients with diarrhea [39].
Cryptosporidium subtypes
All 11 C. parvum isolates were subtyped by sequence analysis of the gp60 gene. However, only five of them produced the expected PCR products, which were identified as two subtypes: IIaA15G2R2 (n = 4, GenBank: KX259142) and IIdA19G1 (n = 1, GenBank: KX259141). IIaA15G2R2 was identical to strains isolated from cattle in the USA (DQ630517), and humans in Canada (DQ192501) and the USA (JX575583), while IIdA19G1 was identical to strains isolated from goats in China (KM199738), cattle in China (HQ009809) and Egypt (JX237824), and humans in China (JQ796092 and JF691561) and Sweden (KU852713). The C. ubiquitum isolates were subtyped to family XIIa and belonged to subtype 2 (GenBank: KX259143-KX259144), sharing 100% homology with a strain isolated from goats in China (KM199742) and a human-derived isolate from Turkey (JX412919). To the best of our knowledge, this is the first time IIaA15G2R2 and IIdA19G1 have been detected in deer.
Giardia assemblage
A total of five G. duodenalis isolates were successfully amplified and sequenced at the SSU rRNA (n = 5), bg (n = 3) and gdh (n = 2) loci, but amplifying failed at the tpi locus despite repeated attempts at molecular analysis using different primers. Sequence analysis showed that all the isolates belonged to G. duodenalis assemblage E. Comparison with SSU rRNA sequences available on GenBank showed 100% sequence identity with sequences of isolates previously recognized from calves (KT922263) and lambs (KT922264) in Ethiopia. Phylogenetic relationship analysis identified G. duodenalis assemblage E (Fig. 2). Two different subtypes were identified at the bg locus, which showed 100% similarity to strains isolated from sheep (GQ337972) in Norway and from calves (KT922247) in Ethiopia, respectively. At the gdh locus, the assemblage E shared 99% similarity with a yak isolate (KP334146) in China.
Discussion
Results obtained from the 662 fecal specimens by both microscopic examination and PCR concurred. PCR is a tool of high sensitivity and specificity [40], particularly for identifying morphologically indistinguishable parasites such as species of Cryptosporidium and assemblages of Giardia, and for detecting their genetic variation. In contrast, Sheather’s sugar flotation technique and Lugol’s iodine staining are routine diagnostics. All the PCR-positive samples found positive by microscopy, and vice versa, may be due to sufficient oocyst and cyst concentrations in fecal specimens.
The Cryptosporidium prevalence of 6.8% in this study was close to the prevalence of 7.84% (25/319, 95% CI: 4.9–10.8%) reported in a study in Japan [9], but higher than that detected in Zhengzhou, China (1.61%, 2/124, 95% CI: 0–3.9%) [14]. It is difficult to explain the discrepancies in the prevalences of Cryptosporidium spp. among different studies because prevalences are affected by many factors, including the age distributions in the sample populations, sample sizes, management systems, seasons, examination methods and ecological conditions.
Studies of Cryptosporidium spp. in cervids have been conducted in several countries and 11 Cryptosporidium species/genotypes have been detected (Table 2). In this study, Cryptosporidium deer genotype was the most frequently detected. Small numbers of the Cryptosporidium deer genotype have been detected in red and roe deer in the UK [12, 15] and in white-tailed deer in the Czech Republic [10], whereas in sika deer in Japan [9] and white-tailed deer in the USA [13], Cryptosporidium deer genotype was the only genotype detected. It appears to be a host-adapted genotype, which has so far only been identified in deer.
Cryptosporidium parvum is one of the two most common Cryptosporidium species in humans [41]. We detected 11 C. parvum isolates, making C. parvum the second-largest cause of infection in cervids in this study. Cryptosporidium parvum infections were observed in red deer and roe deer in the UK [15], red deer in the Czech Republic [7], and white-tailed and black-tailed deer in the USA [4, 11]. We identified zoonotic subtypes IIaA15G2R2 and IIdA19G1 based on a sequence analysis of the gp60 gene. Thus far, at least 14 C. parvum subtype families (IIa-IIi and IIk-IIo) have been found [2, 42]. IIa is the predominant subtype family in animals and humans worldwide, and IId is another major zoonotic subtype family reported in Europe, Asia, Egypt and Australia [43]. In China, most C. parvum isolates belong to subtype IId, including IIdA15G1 found in rodents, cattle and yaks [44,45,46], IIdA18G1 found in yaks [46], and IIdA19G1 found in cattle, humans, goats, yaks and urban wastewater [46,47,48,49,50,51]. In contrast, only a few IIa isolates have been detected in yaks and goats [48, 52], and IIc has been found in monkeys [53]. Subtype IIaA15G2R2 has previously been reported in humans, calves and water in the USA and Canada [54,55,56]. The above findings that the same gp60 gene sequences of the C. parvum isolates have been found in humans and animals, suggest that deer infected with C. parvum in the areas we investigated might pose a threat to local people and animals by shedding oocysts in their feces, thereby contaminating the environment and food sources.
Cryptosporidium ubiquitum, previously called the Cryptosporidium cervine genotype, has been detected in sika deer in China [14], white-tailed deer in the USA [11], swamp deer in Nepal [5], roe deer in the UK [12], red deer in the Czech Republic [10] and deer in Australia [16]. In this study, three C. ubiquitum isolates were identified as zoonotic XIIa subtype 2, which has been found in domestic and wild ruminants, as well as in humans [34]. Despite its low prevalence in this study, C. ubiquitum may be a pathogen of public health concern in this area given its broad host range including rodents, carnivores, primates, domestic and wild ruminants, as well as humans in the UK, Slovenia, the USA, Canada, Spain and New Zealand [34].
Although Cryptosporidium muris is typically a parasite of mice and rats, it has a wide range of host species, including rodents, cats, marsupials (bilbies) and other mammals [57,58,59]. Cryptosporidium muris has also been identified in human cryptosporidiosis cases in many countries, including Thailand, Iran, India, Indonesia, Saudi Arabia, Kenya, Peru and France [60,61,62,63,64,65,66,67], and found in red and white-tailed deer in the Czech Republic [10]. Cryptosporidium suis naturally infects pigs worldwide, but has also been found in cattle, rodents, humans and chimpanzees [68], whereas the C. suis-like genotype has been reported in cattle and rodents, as well as in humans [12, 69, 70]. In this study, only one C. muris isolate and one C. suis-like isolate were identified. The susceptibility of deer to C. muris and C. suis-like is unclear due to inadequate data on these two Cryptosporidium species in cervids.
Cryptosporidium andersoni is predominantly detected in domestic cattle, although it has occasionally been found in other animals, including Bactrian camels, sheep and goats [71]. Only a few cases of human C. andersoni infection have been reported in France, Malawi, Iran, England and Australia, as well as in China [72]. Studies have reported 34 cases of C. andersoni infection in 252 human patients with diarrhea in Shanghai [73], and 21 out of 232 in Jiangsu Province [74]. To the best of our knowledge, this study is the first time C. andersoni has been confirmed in cervids. Five C. andersoni isolates were identified in deer, sharing 100% homology with strains isolated from outpatients with diarrhea in Jiangsu Province, China (KF826313 and KF826314) [74]. The source of C. andersoni infection and its transmission dynamics need further investigation to elucidate the zoonotic potential of C. andersoni in deer in China.
The prevalence of G. duodenalis was 0.76% in this survey, which is lower than that reported in fallow deer in Italy (11.5%) [21], red deer in Croatia (24.0%) [19], red deer, roe deer and moose in Poland (17.0–22.9%) [28], roe deer, reindeer and moose in Norway (7.1–15.5%) [75] and deer in Spain (5.4–8.9%) [6, 20, 76]. Globally, varying prevalence rates of G. duodenalis have been reported in fallow deer, red deer, roe deer, moose, caribou, reindeer, sambar deer and white-tailed deer in the USA, Canada, Australia, Croatia, Spain, Poland, Italy, the Netherlands, Norway and Sweden, ranging between 0.6–24.0% [6, 13, 16, 19,20,21,22,23,24,25,26,27,28,29, 75,76,77,78] (Table 3). In the PCR assay, all the samples were failed amplifying Giardia tpi region. A possible explanation for the result is that inhibitory problems existed in PCRs.
At the molecular level, this study is the first to characterize G. duodenalis from cervids in China, and only assemblage E was identified. Worldwide, less than 100 G. duodenalis isolates from cervids have been analyzed, among which assemblages A, B, D and E were detected [6, 13, 16, 19,20,21,22,23,24,25,26,27,28,29]. Assemblage A, including sub-assemblages A-I (infecting most animals), A-II (mainly found in humans), and A-III (mainly infecting wild ruminants), have previously been reported from red deer, roe deer, fallow deer, reindeer, white-tailed deer and moose in the USA, Croatia, Spain, Poland, Italy, Norway and Australia [6, 13, 16, 19, 21,22,23,24,25,26,27, 29]. Assemblage A is most frequently found assemblage in cervids, while assemblages B and D have only been found in red and roe deer in eastern Poland and Croatia, respectively [19, 28]. In Sweden assemblage E has been detected in a fallow deer [24]. Giardia duodenalis assemblage E has a wide distribution in domestic mammals, including cattle, water buffaloes, sheep, goats and pigs, and is the predominant assemblage found in these animals in the USA, Europe and Australia [1]. However, assemblage E has rarely been identified in wild hoofed animals, and thus may reflect an adaption to these animals following domestication [19]. Moreover, assemblage E has also been found in NHPs from western Uganda and China [79, 80] and in humans from Egypt, Brazil and Australia, suggesting zoonotic transmission of assemblage E [81,82,83,84,85]. Although assemblage E was detected in low numbers in this study, it is still important to understand the public health risk posed by the Giardia species and assemblages infection in cervids in the region.
Conclusions
The 662 samples collected from red deer, Père David’s deer, and sika deer were screened for the presence of Cryptosporidium and G. duodenalis. C. parvum, C. andersoni, C. ubiquitum, C. muris, Cryptosporidium suis-like genotype, Cryptosporidium deer genotype and G. duodenalis assemblage E were detected in this study. This is the first study to report C. andersoni, as well as C. parvum zoonotic subtypes IIaA15G2R2 and IIdA19G1 in cervids. Deer farming has become an important component of China’s animal breeding industry. Farming increases the potential contact between deer and humans, and intensifies the numbers of animals, which potentially increases the numbers of shed (oo) cysts in the environment. Given that we have detected human pathogens in deer in China, further investigations into the transmission dynamics of these pathogens would be warranted.
Abbreviations
- bg :
-
β-giardin
- gdh :
-
Glutamate dehydrogenase
- gp60 :
-
60 kDa glycoprotein
- PCR:
-
Polymerase chain reaction
- SSU rRNA:
-
Small subunit ribosomal ribonucleic acid
- tpi :
-
Triosephosphate isomerase
References
Feng Y, Xiao L. Zoonotic potential and molecular epidemiology of Giardia species and giardiasis. Clin Microbiol Rev. 2011;24:110–40.
Xiao L. Molecular epidemiology of cryptosporidiosis: an update. Exp Parasitol. 2010;124:80–9.
Thompson RC, Ash A. Molecular epidemiology of Giardia and Cryptosporidium infections. Infect Genet Evol. 2016;40:315–23.
Deng MQ, Cliver DO. Improved immunofluorescence assay for detection of Giardia and Cryptosporidium from asymptomatic adult cervine animals. Parasitol Res. 1999;85:733–6.
Feng Y, Karna SR, Dearen TK, Singh DK, Adhikari LN, Shrestha A, et al. Common occurrence of a unique Cryptosporidium ryanae variant in zebu cattle and water buffaloes in the buffer zone of the Chitwan National Park. Nepal. Vet Parasitol. 2012;185:309–14.
Garcia-Presedo I, Pedraza-Diaz S, Gonzalez-Warleta M, Mezo M, Gomez-Bautista M, Ortega-Mora LM, et al. The first report of Cryptosporidium bovis, C. ryanae and Giardia duodenalis sub-assemblage A-II in roe deer (Capreolus capreolus) in Spain. Vet Parasitol. 2013;197:658–64.
Hajdušek O, Ditrich O, Šlapeta J. Molecular identification of Cryptosporidium spp. in animal and human hosts from the Czech Republic. Vet Parasitol. 2004;122:183–92.
Jellison KL, Lynch AE, Ziemann JM. Source tracking identifies deer and geese as vectors of human-infectious Cryptosporidium genotypes in an urban/suburban watershed. Environ Sci Technol. 2009;43:4267–72.
Kato S, Yanagawa Y, Matsuyama R, Suzuki M, Sugimoto C. Molecular identification of the Cryptosporidium deer genotype in the Hokkaido sika deer (Cervus nippon yesoensis) in Hokkaido. Japan. Parasitol Res. 2016;115:1463–71.
Kotková M, Němejc K, Sak B, Hanzal V, Květoňová D, Hlásková L, et al. Cryptosporidium ubiquitum, C. muris and Cryptosporidium deer genotype in wild cervids and caprines in the Czech Republic. Folia Parasitol. 2016;63:2016.003.
Perz JF, Le Blancq SM. Cryptosporidium parvum infection involving novel genotypes in wildlife from lower New York State. Appl Environ Microbiol. 2001;67:1154–62.
Robinson G, Chalmers RM, Stapleton C, Palmer SR, Watkins J, Francis C, et al. A whole water catchment approach to investigating the origin and distribution of Cryptosporidium species. J Appl Microbiol. 2011;111:717–30.
Santin M, Fayer R. Enterocytozoon bieneusi, Giardia, and Cryptosporidium infecting white-tailed deer. J Eukaryot Microbiol. 2015;62:34–43.
Wang R, Wang J, Sun M, Dang H, Feng Y, Ning C, et al. Molecular characterization of the Cryptosporidium cervine genotype from a sika deer (Cervus nippon Temminck) in Zhengzhou, China and literature review. Parasitol Res. 2008;103:865–9.
Wells B, Shaw H, Hotchkiss E, Gilray J, Ayton R, Green J, et al. Prevalence, species identification and genotyping Cryptosporidium from livestock and deer in a catchment in the Cairngorms with a history of a contaminated public water supply. Parasit Vectors. 2015;8:66.
Koehler AV, Haydon SR, Jex AR, Gasser RB. Cryptosporidium and Giardia taxa in faecal samples from animals in catchments supplying the city of Melbourne with drinking water (2011 to 2015). Parasit Vectors. 2016;9:315.
Xiao L, Sulaiman IM, Ryan UM, Zhou L, Atwill ER, Tischler ML, et al. Host adaptation and host-parasite co-evolution in Cryptosporidium: implications for taxonomy and public health. Int J Parasitol. 2002;32:1773–85.
Kváč M, McEvoy J, Stenger B, Clark M. Cryptosporidiosis in other vertebrates. In: Cacciò SM, Widmer G, editors. Cryptosporidium: parasite and disease. Vienna: Springer; 2014. p. 237–323.
Beck R, Sprong H, Lucinger S, Pozio E, Caccio SM. A large survey of Croatian wild mammals for Giardia duodenalis reveals a low prevalence and limited zoonotic potential. Vector Borne Zoonotic Dis. 2011;11:1049–55.
Castro-Hermida JA, Garcia-Presedo I, Gonzalez-Warleta M, Mezo M. Prevalence of Cryptosporidium and Giardia in roe deer (Capreolus capreolus) and wild boars (Sus scrofa) in Galicia (NW Spain). Vet Parasitol. 2011;179:216–9.
Lalle M, Frangipane di Regalbono A, Poppi L, Nobili G, Tonanzi D, Pozio E, et al. A novel Giardia duodenalis assemblage A subtype in fallow deer. J Parasitol. 2007;93:426–8.
Solarczyk P, Majewska AC, Moskwa B, Cabaj W, Dabert M, Nowosad P. Multilocus genotyping of Giardia duodenalis isolates from red deer (Cervus elaphus) and roe deer (Capreolus capreolus) from Poland. Folia Parasitol. 2012;59:237–40.
Trout JM, Santin M, Fayer R. Identification of assemblage A Giardia in white-tailed deer. J Parasitol. 2003;89:1254–5.
Lebbad M, Mattsson JG, Christensson B, Ljungstrom B, Backhans A, Andersson JO, et al. From mouse to moose: multilocus genotyping of Giardia isolates from various animal species. Vet Parasitol. 2010;168:231–9.
Caccio SM, Beck R, Lalle M, Marinculic A, Pozio E. Multilocus genotyping of Giardia duodenalis reveals striking differences between assemblages A and B. Int J Parasitol. 2008;38:1523–31.
Robertson LJ, Forberg T, Hermansen L, Hamnes IS, Gjerde B. Giardia duodenalis cysts isolated from wild moose and reindeer in Norway: genetic characterization by PCR-rflp and sequence analysis at two genes. J Wildl Dis. 2007;43:576–85.
Miska KB, Jenkins MC, Trout JM, Santin M, Fayer R. Detection and comparison of Giardia virus (Glv) from different assemblages of Giardia duodenalis. J Parasitol. 2009;95:1197–200.
Stojecki K, Sroka J, Caccio SM, Cencek T, Dutkiewicz J, Kusyk P. Prevalence and molecular typing of Giardia duodenalis in wildlife from eastern Poland. Folia Parasitol. 2015;62:42.
Van der Giessen JWB, de Vries A, Roos M, Wielinga P, Kortbeek LM, Mank TG. Genotyping of Giardia in Dutch patients and animals: a phylogenetic analysis of human and animal isolates. Int J Parasitol. 2006;36:849–58.
Ohtaishi N, Gao Y. A review of the distribution of all species of deer (Tragulidae, Moschidae and Cervidae) in China. Mammal Review. 1990;20:125–44.
Li ZP, Liu HL, Li GY, Bao K, Wang KY, Xu C, et al. Molecular diversity of rumen bacterial communities from tannin-rich and fiber-rich forage fed domestic sika deer (Cervus nippon) in China. BMC Microbiol. 2013;13:151.
Cao K. Research on the mi-deer. Shanghai: Shanghai Scientific Education Publishing House; 2005. p. 246.
Jiang JL, Alderisio KA, Xiao LH. Distribution of Cryptosporidium genotypes in storm event water samples from three watersheds in New York. Appl Environ Microbiol. 2005;71:4446–54.
Li N, Xiao LH, Alderisio K, Elwin K, Cebelinski E, Chalmers R, et al. Subtyping Cryptosporidium ubiquitum, a zoonotic pathogen emerging in humans. Emerg Infect Dis. 2014;20:217–24.
Wang HY, Zhao GH, Chen GY, Jian FC, Zhang SM, Feng C, et al. Multilocus genotyping of Giardia duodenalis in dairy cattle in Henan. China. PLoS One. 2014;9(6):e100453.
Sulaiman IM, Fayer R, Bern C, Gilman RH, Trout JM, Schantz PM, et al. Triosephosphate isomerase gene characterization and potential zoonotic transmission of Giardia duodenalis. Emerg Infect Dis. 2003;9:1444–52.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Amer S, Honma H, Ikarashi M, Tada C, Fukuda Y, Suyama Y, et al. Cryptosporidium genotypes and subtypes in dairy calves in Egypt. Vet Parasitol. 2010;169:382–6.
Jiang YY, Ren JH, Yuan ZY, Liu AQ, Zhao H, Liu H, et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province. China. BMC Infect Dis. 2014;14:555.
Fayer R, Santin M, Macarisin D. Detection of concurrent infection of dairy cattle with Blastocystis, Cryptosporidium, Giardia, and Enterocytozoon by molecular and microscopic methods. Parasitol Res. 2012;111:1349–55.
Xiao L, Fayer R. Molecular characterisation of species and genotypes of Cryptosporidium and Giardia and assessment of zoonotic transmission. Int J Parasitol. 2008;38:1239–55.
Insulander M, Silverlas C, Lebbad M, Karlsson L, Mattsson JG, Svenungsson B. Molecular epidemiology and clinical manifestations of human cryptosporidiosis in Sweden. Epidemiol Infect. 2013;141:1009–20.
Wang R, Zhang L, Axen C, Bjorkman C, Jian F, Amer S, et al. Cryptosporidium parvum IId family: clonal population and dispersal from western Asia to other geographical regions. Sci Rep. 2014;4:4208.
Cui ZH, Wang RJ, Huang JY, Wang HY, Zhao JF, Luo NN, et al. Cryptosporidiosis caused by Cryptosporidium parvum subtype IIdA15G1 at a dairy farm in northwestern China. Parasit Vectors. 2014;7:529.
Huang JY, Yue DY, Qi M, Wang RJ, Zhao JF, Li JQ, et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in dairy cattle in Ningxia, northwestern China. BMC Vet Res. 2014;10:292.
Qi M, Cai J, Wang R, Li J, Jian F, Huang J, et al. Molecular characterization of Cryptosporidium spp. and Giardia duodenalis from yaks in the central western region of China. BMC Microbiol. 2015;15:108.
Li N, Xiao L, Wang L, Zhao S, Zhao X, Duan L, et al. Molecular surveillance of Cryptosporidium spp., Giardia duodenalis, and Enterocytozoon bieneusi by genotyping and subtyping parasites in wastewater. PLoS Negl Trop Dis. 2012;6:e1809.
Mi R, Wang X, Huang Y, Zhou P, Liu Y, Chen Y, et al. Prevalence and molecular characterization of Cryptosporidium in goats across four provincial level areas in China. PLoS One. 2014;9:e111164.
Wang L, Zhang H, Zhao X, Zhang L, Zhang G, Guo M, et al. Zoonotic Cryptosporidium species and Enterocytozoon bieneusi genotypes in HIV-positive patients on antiretroviral therapy. J Clin Microbiol. 2013;51:557–63.
Wang R, Wang H, Sun Y, Zhang L, Jian F, Qi M, et al. Characteristics of Cryptosporidium transmission in preweaned dairy cattle in Henan. China. J Clin Microbiol. 2011;49:1077–82.
Zhang W, Wang R, Yang F, Zhang L, Cao J, Zhang X, et al. Distribution and genetic characterizations of Cryptosporidium spp. in pre-weaned dairy calves in Northeastern China's Heilongjiang Province. PLoS One. 2013;8:e54857.
Ma J, Cai J, Ma J, Feng Y, Xiao L. Occurrence and molecular characterization of Cryptosporidium spp. in yaks (Bos grunniens) in China. Vet Parasitol. 2014;202:113–8.
Ye J, Xiao L, Ma J, Guo M, Liu L, Feng Y. Anthroponotic enteric parasites in monkeys in public park, China. Emerg Infect Dis. 2012;18:1640–3.
Herges GR, Widmer G, Clark ME, Khan E, Giddings CW, Brewer M, et al. Evidence that Cryptosporidium parvum populations are panmictic and unstructured in the upper midwest of the United States. Appl Environ Microbiol. 2012;78:8096–101.
Trotz-Williams LA, Martin DS, Gatei W, Cama V, Peregrine AS, Martin SW, et al. Genotype and subtype analyses of Cryptosporidium isolates from dairy calves and humans in Ontario. Parasitol Res. 2006;99:346–52.
Xiao LH, Zhou L, Santin M, Yang WL, Fayer R. Distribution of Cryptosporidium parvum subtypes in calves in eastern United States. Parasitol Res. 2007;100:701–6.
Chappell CL, Okhuysen PC, Langer-Curry RC, Lupo PJ, Widmer G, Tzipori S. Cryptosporidium muris: infectivity and illness in healthy adult volunteers. Am J Trop Med Hyg. 2015;92:50–5.
Ryan U, Fayer R, Xiao LH. Cryptosporidium species in humans and animals: current understanding and research needs. Parasitology. 2014;141:1667–85.
Yang RC, Ying JLJ, Monis P, Ryan U. Molecular characterisation of Cryptosporidium and Giardia in cats (Felis catus) in western Australia. Exp Parasitol. 2015;155:13–8.
Al-Brikan FA, Salem HS, Beeching N, Hilal N. Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J Egypt Soc Parasitol. 2008;38:645–58.
Azami M, Moghaddam DD, Salehi R, Salehi M. The identification of Cryptosporidium species (Protozoa) in Ifsahan, Iran by PCR-RFLP analysis of the 18S rRNA gene. Mol Biol. 2007;41:934–9. (In Russian)
Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, et al. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg. 2006;75:78–82.
Guyot K, Follet-Dumoulin A, Lelievre E, Sarfati C, Rabodonirina M, Nevez G, et al. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–80.
Katsumata T, Hosea D, Ranuh IG, Uga S, Yanagi T, Kohno S. Short report: possible Cryptosporidium muris infection in humans. Am J Trop Med Hyg. 2000;62:70–2.
Muthusamy D, Rao SS, Ramani S, Monica B, Banerjee I, Abraham OC, et al. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J Clin Microbiol. 2006;44:632–4.
Palmer CJ, Xiao L, Terashima A, Guerra H, Gotuzzo E, Saldias G, et al. Cryptosporidium muris, a rodent pathogen, recovered from a human in Peru. Emerg Infect Dis. 2003;9:1174–6.
Tiangtip R, Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health. 2002;7:357–64.
Zahedi A, Paparini A, Jian F, Robertson I, Ryan U. Public health significance of zoonotic Cryptosporidium species in wildlife: critical insights into better drinking water management. Int J Parasitol Parasites Wildl. 2015;5(1):88–109.
Fayer R, Santin M, Trout JM, Greiner E. Prevalence of species and genotypes of Cryptosporidium found in 1-2-year-old dairy cattle in the eastern United States. Vet Parasitol. 2006;135:105–12.
Ng-Hublin JS, Singleton GR, Ryan U. Molecular characterization of Cryptosporidium spp. from wild rats and mice from rural communities in the Philippines. Infect Genet Evol. 2013;16:5–12.
Feng Y, Yang W, Ryan U, Zhang L, Kvac M, Koudela B, et al. Development of a multilocus sequence tool for typing Cryptosporidium muris and Cryptosporidium andersoni. J Clin Microbiol. 2011;49:34–41.
Nichols GL, Chalmers RM, Hadfield SJ. Molecular epidemiology of human cryptosporidiosis. In: Cacciò SM, Widmer G, editors. Cryptosporidium: parasite and disease. Vienna: Springer; 2014. p. 81–147.
Liu H, Shen Y, Yin J, Yuan Z, Jiang Y, Xu Y, et al. Prevalence and genetic characterization of Cryptosporidium, Enterocytozoon, Giardia and Cyclospora in diarrheal outpatients in China. BMC Infect Dis. 2014;14:25.
Jiang Y, Ren J, Yuan Z, Liu A, Zhao H, Liu H, et al. Cryptosporidium andersoni as a novel predominant Cryptosporidium species in outpatients with diarrhea in Jiangsu Province, China. BMC Infect Dis. 2014;14:555.
Hamnes IS, Gjerde B, Robertson L, Vikoren T, Handeland K. Prevalence of Cryptosporidium and Giardia in free-ranging wild cervids in Norway. Vet Parasitol. 2006;141:30–41.
Castro-Hermida JA, Garcia-Presedo I, Almeida A, Gonzalez-Warleta M, Da Costa JMC, Mezo M. Cryptosporidium spp. and Giardia duodenalis in two areas of Galicia (NW Spain). Sci Total Environ. 2011;409:2451–9.
Johnson D, Harms NJ, Larter NC, Elkin BT, Tabel H, Wei G. Serum biochemistry, serology, and parasitology of boreal caribou (Rangifer tarandus caribou) in the Northwest Territories, Canada. J Wildl Dis. 2010;46:1096–107.
Rickard LG, Siefker C, Boyle CR, Gentz EJ. The prevalence of Cryptosporidium and Giardia spp. in fecal samples from free-ranging white-tailed deer (Odocoileus virginianus) in the southeastern United States. J Vet Diagn Invest. 1999;11:65–72.
Du SZ, Zhao GH, Shao JF, Fang YQ, Tian GR, Zhang LX, et al. Cryptosporidium spp., Giardia intestinalis, and Enterocytozoon bieneusi in captive non-human primates in Qinling Mountains. Korean J Parasitol. 2015;53:395–402.
Johnston AR, Gillespie TR, Rwego IB, McLachlan TL, Kent AD, Goldberg TL. Molecular epidemiology of cross-species Giardia duodenalis transmission in western Uganda. PLoS Negl Trop Dis. 2010;4:e683.
Abdel-Moein KA, Saeed H. The zoonotic potential of Giardia intestinalis assemblage E in rural settings. Parasitol Res. 2016;115:3197–202.
Fantinatti M, Bello AR, Fernandes O, Da-Cruz AM. Identification of Giardia lamblia assemblage E in humans points to a new anthropozoonotic cycle. J Infect Dis. 2016;214:1256–9.
Foronda P, Bargues MD, Abreu-Acosta N, Periago MV, Valero MA, Valladares B, et al. Identification of genotypes of Giardia intestinalis of human isolates in Egypt. Parasitol Res. 2008;103:1177–81.
Helmy YA, Klotz C, Wilking H, Krucken J, Nockler K, Von Samson-Himmelstjerna G, et al. Epidemiology of Giardia duodenalis infection in ruminant livestock and children in the Ismailia Province of Egypt: insights by genetic characterization. Parasit Vectors. 2014;7:321.
Zahedi A, Field D, Ryan U. Molecular typing of Giardia duodenalis in humans in Queensland - first report of Assemblage E. Parasitology. 2017;144:1154–61.
Acknowledgments
We thank Dr Yongjun Wen, the associate research fellow of Institute of Special Economic Animal and Plant Science, Chinese Academy of Agricultural Sciences, for his valuable assistance in sample collection. We thank Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Funding
This study was supported in part by the Key Program of the National Natural Science Foundation of China (31330079), the National Natural Science Foundation of China (313020793, 31110103901), and the Program for Science & Technology Innovation Talents in Universities of Henan Province (16HASTIT018).
Availability of data and materials
All data generated or analyzed during this study are included in this published article. The sequences are submitted in the GenBank database under accession numbers KX259127-KX259145 and MG921620-MG921622.
Author information
Authors and Affiliations
Contributions
RJW and LXZ conceived and designed the experiments. JYH, ZJZ and YY performed the experiments. JFZ, FCJ, WYZ and CSN analyzed the data. JYH, RJW and LXZ wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This study was conducted in accordance with the Chinese Laboratory Animal Administration Act of 1988. The research protocol was reviewed and approved by the Research Ethics Committee of Henan Agricultural University. Permission was obtained from all farm owners before the fecal samples were collected.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Huang, J., Zhang, Z., Zhang, Y. et al. Prevalence and molecular characterization of Cryptosporidium spp. and Giardia duodenalis in deer in Henan and Jilin, China. Parasites Vectors 11, 239 (2018). https://doi.org/10.1186/s13071-018-2813-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13071-018-2813-9