Abstract
Background
In B-cell precursor acute lymphoblastic leukemia (BCP-ALL) PAX5, a transcription factor pivotal for B-cell commitment and maintenance, is frequently affected by genetic alterations. In 2-3 % of the cases PAX5 rearrangements result in the expression of oncogenic fusion genes. The encoded chimeric proteins consist of the N-terminal PAX5 DNA-binding paired domain, which is fused to the C-terminal domains of a remarkable heterogeneous group of partner proteins.
Results
Employing fluorescence in situ hybridization and molecular methods PAX5-KIAA1549L was identified as novel fusion gene in a case of pediatric BCP-ALL.
Conclusion
Our report underlines the high diversity of PAX5 fusion partners in BCP-ALL and we describe the second involvement of KIAA1549L in a genetic rearrangement in acute leukemia.
Similar content being viewed by others
Background
Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy and one of the leading causes of cancer-related death in children and young adults. ALL is characterized by a plethora of somatic mutations, which are involved in the pathogenesis and progression of the disease [1]. In this regard, in childhood B-cell precursor ALL (BCP-ALL) PAX5, a transcription factor crucial for B-cell commitment and maintenance [2], is frequently affected by genetic alterations comprising deletions, point mutations, amplifications, and chromosomal rearrangements [3–7]. The latter occur in 2-3 % of the cases and result in the expression of chimeric proteins consisting of the PAX5 DNA-binding paired domain (PD) at the N-terminus, fused to the C-terminal domains of a highly diverse group of partner proteins [3–5, 8]. A multitude of functionally different PAX5 partners, including transcription factors, chromatin regulators, protein kinases, and structural proteins, have been identified and to date more than 20 in-frame PAX5 fusions have been described [3–5, 8–15]. The unifying feature of all PAX5 chimeras, which are generally suggested to act as aberrant transcription factors antagonizing wild-type PAX5 function, is the retention of the DNA-binding domain [2, 3, 9, 16–18]. However, the partner protein appears to modulate the function of the different PAX5 fusion proteins, suggesting differences in the development of the respective leukemia [19, 20]. In this work we describe a novel PAX5 rearrangement in a case of pediatric BCP-ALL in which PAX5 is fused to KIAA1549L.
Case presentation
A 2.2-year old girl was diagnosed with BCP-ALL. The bone marrow and the peripheral blood showed an infiltration with 94 % and 89 % blast cells of L1 morphology, respectively. Immunophenotyping demonstrated CD19+, CD10+, cyCD22+, HLA-DR+, TdT+, cyIgM+, mIgM-, MPO-, CD34-, CD14-, CD13-, CD33-, cyCD3-, and CD7- blast cells with coexpression of the myeloid marker CDw65 on 20 % of them. Cytogenetic and molecular analyses showed a normal 46,XX[20] karyotype and negativity for the ETV6-RUNX1, KMT2A-AFF1 (alias MLL-AF4), BCR-ABL1, and TCF3-PBX1 fusion genes.
The patient was enrolled in the ALL-BFM 95 study (ClinicalTrials.gov Identifier: NCT00411541) and treated in the intermediate risk arm of the protocol [21, 22]. The patient was randomized into the methotrexate and cytarabine (MCA) protocol, which included an intensification of the extracompartement phase with an intermediate dose cytarabine in addition to high-dose methotrexate. For maintenance therapy the patient was randomized into the treatment arm, which received an intensified therapy with additional pulses of dexamethasone and vincristine. The patient is in long-term remission 8.6 years after initial diagnosis.
Material and methods
Cytogenetic and fluorescence in situ hybridization (FISH) analysis
Cytogenetic and FISH analyses were performed according to standard techniques. FISH was conducted using PAX5 exon-specific cosmid probes cos-hPAX5-1 (exons 2–5) and cos-hPAX5-3 (exons 9-10) as previously described [4].
Rapid amplification of cDNA ends (RACE) and reverse transcription-polymerase chain reaction (RT-PCR)
RNA isolation was performed with the Qiagen RNeasy Mini-Kit (Qiagen, Germany) following the manufacturer’s instructions. RACE ready cDNA was generated using 220 ng total RNA input and the SMARTer™ RACE cDNA Amplification Kit (Clontech/Takara, France). 3′-RACE-PCR and 3′-nested RACE-PCR were performed with the Advantage 2 PCR Kit (Clontech/Takara, France) and standard PCR reagents (Sigma, Austria), respectively.
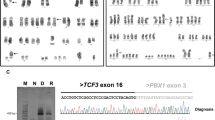
RT-PCR for the detection of PAX5-KIAA1549L transcripts was performed according to standard procedures using primers located in exon 5 of PAX5 (PAX5ex5_F1 5′-TACTCCATCAGCGGCATCC-3′) and exon 20 of KIAA1549L (KIAA1549Lex20_R2: 5′-AACACGTAGGCATGGGAAAC-3′). Amplification products were directly sequenced (Microsynth AG, Austria) and sequence analysis was conducted using the CLC Main Workbench 6.0 (CLC bio, Denmark).
Reference sequences, exon nomenclature, and protein analysis
The exon nomenclature used corresponds to that of the NCBI database (accessed December 2014) and the reference sequences NM_016734.2 for PAX5 and NM_012194 for KIAA1549L were used. Protein motif search was performed with Psort II Prediction [23] and NCBI_Protein [24]. The conservation of KIAA1549L across species was derived from NCBI_HomoloGene [25].
Detection of copy number alterations (CNAs)
Genome-wide CNAs were determined using the CytoScan HD array platform (Affymetrix, USA). This array platform contains 2.67 million probes, including 1.9 million copy number probes and 0.75 million SNP probes. Array analysis and interpretation was conducted with the Chromosome Analysis Suite (ChAS 2.0.0) software (Affymetrix, USA). Genome segment filters were set to a marker count of 50 and a size of 50 kb for gains and losses. The ChAS Browser Annotations version NetAffx built 32.3 (UCSC genome assembly hg19) was used for analysis.
Results and discussion
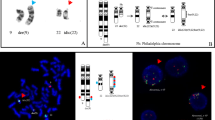
FISH analysis of the diagnostic bone marrow using PAX5-specific probes showed a deletion of the PAX5 3’-end in 72 % of the interphase cells, which is highly suggestive of the presence of a PAX5 fusion gene (Fig. 1a). 3’-RACE-PCR and subsequent verification by RT-PCR identified KIAA1549L (alias C11orf41), located at 11p13, as novel PAX5 fusion partner (Fig. 1b). Sequence analysis revealed an in-frame PAX5-KIAA1549L fusion transcript in which exon 6 of PAX5 is fused to exon 16 of KIAA1549L (Fig. 1c). The predicted fusion protein retains the DNA-binding paired domain (PD), the octapeptide motif, the nuclear localization signal, and the homeodomain of PAX5, which are fused to the C-terminus of KIAA1549L (Fig. 1d).
Cytogenetic and molecular analysis of PAX5-KIAA1549L. a Interphase FISH using PAX5 exon-specific cosmid probes cos-hPAX5-1 (exons 2–5, red signals) and cos-hPAX5-3 (exons 9–10, green signals) showing loss of a PAX5 3’-end. b RT-PCR using primers specific for PAX5 exon 5 and KIAA1549L exon 20 resulting in amplification of PAX5-KIAA1549L fusion transcripts. Sequencing of the second smaller band, indicated by an asterisk (*), revealed a splice variant of PAX5-KIAA1549L, lacking exon 16 of KIAA1549L. M, molecular weight marker DNA ladder-mix (Peqlab); lane 1, patient; lane 2, normal control; lane 3, water control. c Sequence analysis of the predominant RT-PCR product showing the presence of an in-frame fusion between PAX5 exon 6 and KIAA1549L exon 16. d Schematic representation of the PAX5 wild-type and the PAX5-KIAA1549L proteins. PD, paired domain; 8, octapeptide; HD, partial homeodomain; TA, transactivation domain; ID, inhibitory domain; arrows indicate the nuclear localization signal of PAX5; aa, amino acids. e Chromosomes 9 and 11 with areas of genetic loss (red) detected by the CytoScan HD array and selected genes included in these areas. f Detailed view of PAX5 on chromosome 9 (left panel) and KIAA1549L on chromosome 11 (right panel). The blue dashed lines indicate the breakpoints and the transitions from areas with normal and deleted (red bars) copy number states. The retained exons 1–6 of PAX5 and exons 16–20 of KIAA1549L correspond to the fusion gene detected by RT-PCR
KIAA1549L is a 1849 amino acids long protein with highly conserved regions whose functions have not been determined yet. As predicted by protein motif search, KIAA1549L harbors only a potential transmembrane domain encoded by exon 10, which, however, is not retained in the PAX5-KIAA1549L chimera. KIAA1549L is preferentially expressed in the human cerebral cortex and immunohistochemistry data suggest that KIAA1549L is localized in the nucleus as well as in cytoplasmatic vesicles [26, 27].
The opposite orientation of the partner genes – PAX5 centromere-telomere and KIAA1549L telomere-centromere – excludes the formation of the PAX5-KIAA1549L fusion by a simple reciprocal translocation and their location and orientation suggested the presence of a dic(9;11) chromosome. Since the lack of metaphases precluded a detailed FISH analysis of the chromosomes involved in the rearrangement, we analyzed the genomic DNA using the CytoScan HD array, which – apart from the genome-wide detection of copy number alterations (CNAs) – permits to uncover unbalanced translocations. However, in contrast to PAX5-ETV6 and PAX5-C20orf112, which are generated by the formation of dic(9;12) and dic(9;20) chromosomes, respectively [12, 28], in case of PAX5-KIAA1549L the CNAs did not corroborate the presence of a dic(9;11). Instead of losses of almost the entire p-arms of chromosomes 9 and 11, which would reflect the presence of a dic(9;11), we observed only minor deletions on the involved chromosomes (Fig. 1e). Consequently, similarly to other PAX5 fusions [4, 8], also the PAX5-KIAA1549L fusion is most likely generated by a complex rearrangement.
A more detailed analysis of the CNAs of chromosomes 9 and 11 revealed a 324 kb heterozygous deletion on chromosome 9, which – amongst other genes – affected PAX5 exons 7–10, and a 578 kb heterozygous deletion on chromosome 11, including KIAA1549L exons 1–15 (Fig. 1f). In line with the molecular analysis of the PAX5-KIAA1549L fusion gene (Fig. 1c), PAX5 exons 1–6 and KIAA1549L exons 16–20, which are present in the fusion gene, were retained (Fig. 1f). These data indicate that only PAX5-KIAA1549L but no reciprocal transcripts are expressed, which supports the previous finding that in most PAX5-rearranged cases only PAX5 fusion transcripts but no corresponding reciprocal ones are detectable [4, 8, 29].
With regard to other CNAs and in line with previous findings in PAX5-rearranged leukemia [8], we generally detected only a few minor gains and losses (Additional file 1). Chromosome 9 showed an additional heterozygous loss of a 4760 kb region at 9p21, within which we observed a smaller homozygous deletion affecting CDKN2A/B (Fig. 1e, Fig. 2a, Additional file 1). CDKN2A/B is commonly deleted in ALL [30] and its hetero- or homozygous loss is also frequently observed in dic(9;20) leukemia [31–33], which in some cases expresses the PAX5-C20orf112 fusion gene [12, 32, 34, 35]. Furthermore, formation of the dic(9;12) resulting in the expression of the PAX5-ETV6 fusion is invariably associated with a heterozygous deletion of CDKN2A/B as well [28, 35]. Therefore, loss of CDKN2A/B may represent a secondary genetic lesion that cooperates at least with a subset of PAX5 fusions in the development of BCP-ALL.
Detailed view of gains and losses of chromosomes 9, 11, and X. a Chromosome 9. The heterozygous 4760 kb deletion includes a homozygous deletion of CDKN2A/B indicated by the decrease of the smooth signal to zero. The smaller heterozygous loss affects, amongst other genes, the 3’-end of PAX5. b Chromosome 11. The heterozygous deletion at 11p13 includes the 5’-end of KIAA1549L. c Chromosome X. The changes of the log2 ratios along the chromosome indicate a trisomy of the entire q-arm and a small part of the p-arm as well as loss of most of the p-arm, indicating the presence of an idic(X)(p11)
Remarkably, chromosome X displayed a heterozygous loss of almost an entire p-arm and a gain of one copy of the entire q-arm, indicating the presence of an isodicentric idic(X)(p11) chromosome (Fig. 2c, Additional file 1). While idic(X)(q13) chromosomes are recurrently found in myeloid malignancies [36], an idic(X)(p11) is an extremely rare chromosome abnormality, which according to the Mitelman database [37] has only been described in a single case of Burkitt lymphoma [38].
Since PAX5 fusions in ALL are rather rare [4, 5], their prognostic relevance per se and also the impact of secondary alterations including loss of CDKN2A/B remain to be determined. However, in this context it appears that PAX5 deletions and mutations are not associated with clinical outcome [39] and that the prognostic value of CDKNA/B deletions is cytogenetic subtype and copy number alteration profile dependent [30, 40].
Involvement of KIAA1549L in another chromosome translocation has been previously reported in an adult case of acute myeloid leukemia with a t(11;21)(p14;q22) resulting in a RUNX1-KIAA1549L fusion [41]. In contrast to the RUNX1-KIAA1549L fusion, in which alternatively spliced RUNX1 sequences are fused to exon 13 of KIAA1549L, PAX5 exon 6 is fused to exon 16, indicating different genomic breakpoints in introns 12 and 15 of KIAA1549L, respectively. Furthermore, KIAA1549, a homolog of KIAA1549L, is involved in the recurrent KIAA1549-BRAF fusion gene found in pediatric and adult pilocytic astrocytoma [42].
In summary, we here describe a novel PAX5 fusion partner and the second involvement of KIAA1549L in a leukemic fusion gene, indicating that it may play a so far underestimated role in the development of acute leukemia.
Conclusion
In this study we identified KIAA1549L as novel PAX5 fusion partner, which, on the one hand, increases the number of PAX5 fusion partners found in BCP-ALL, and, on the other hand, shows that KIAA1549L is involved in at least two different fusion genes, emphasizing its potential role in the pathogenesis of acute leukemia.
Consent
The patient was enrolled and treated with informed consent in the ALL-BFM 95 study (ClinicalTrials.gov Identifier: NCT00411541). This study was exclusively performed on material obtained for diagnostic purposes and neither any additional medical intervention nor patient recruitment was necessary.
References
Mullighan CG. Genomic characterization of childhood acute lymphoblastic leukemia. Semin Hematol. 2013;50:314–24.
Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Adv Immunol. 2011;111:179–206.
Mullighan CG, Goorha S, Radtke I, Miller CB, Coustan-Smith E, Dalton JD, et al. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 2007;446:758–64.
Nebral K, Denk D, Attarbaschi A, Konig M, Mann G, Haas OA, et al. Incidence and diversity of PAX5 fusion genes in childhood acute lymphoblastic leukemia. Leukemia. 2009;23:134–43.
Coyaud E, Struski S, Prade N, Familiades J, Eichner R, Quelen C, et al. Wide diversity of PAX5 alterations in B-ALL: a Groupe Francophone de Cytogenetique Hematologique study. Blood. 2010;115:3089–97.
Familiades J, Bousquet M, Lafage-Pochitaloff M, Bene MC, Beldjord K, De Vos J, et al. PAX5 mutations occur frequently in adult B-cell progenitor acute lymphoblastic leukemia and PAX5 haploinsufficiency is associated with BCR-ABL1 and TCF3-PBX1 fusion genes: a GRAALL study. Leukemia. 2009;23:1989–98.
Ofverholm I, Tran AN, Heyman M, Zachariadis V, Nordenskjold M, Nordgren A, et al. Impact of IKZF1 deletions and PAX5 amplifications in pediatric B-cell precursor ALL treated according to NOPHO protocols. Leukemia. 2013;27:1936–9.
Fazio G, Daniele G, Cazzaniga V, Impera L, Severgnini M, Iacobucci I, et al. Three novel fusion transcripts of the paired box 5 gene in B-cell precursor acute lymphoblastic leukemia. Haematologica. 2014. doi:10.3324/haematol.2014.112193.
Bousquet M, Broccardo C, Quelen C, Meggetto F, Kuhlein E, Delsol G, et al. A novel PAX5-ELN fusion protein identified in B-cell acute lymphoblastic leukemia acts as a dominant negative on wild-type PAX5. Blood. 2007;109:3417–23.
Nebral K, Konig M, Harder L, Siebert R, Haas OA, Strehl S. Identification of PML as novel PAX5 fusion partner in childhood acute lymphoblastic leukaemia. Br J Haematol. 2007;139:269–74.
Cazzaniga G, Daniotti M, Tosi S, Giudici G, Aloisi A, Pogliani E, et al. The paired box domain gene PAX5 is fused to ETV6/TEL in an acute lymphoblastic leukemia case. Cancer Res. 2001;61:4666–70.
Kawamata N, Ogawa S, Zimmermann M, Niebuhr B, Stocking C, Sanada M, et al. Cloning of genes involved in chromosomal translocations by high-resolution single nucleotide polymorphism genomic microarray. Proc Natl Acad Sci U S A. 2008;105:11921–6.
Lee ST, Ji Y, Kim HJ, Ki CS, Jung CW, Kim JW, et al. Sequential array comparative genomic hybridization analysis identifies copy number changes during blastic transformation of chronic myeloid leukemia. Leuk Res. 2012;36:418–21.
Holmfeldt L. Genomic- and Transcriptomic Profiling Of Acute Lymphoblastic Leukemia With Dicentric Chromosomes ASH Annual Meeting Abstracts. Blood. 2013;122:234.
Roberts KG, Li Y, Payne-Turner D, Harvey RC, Yang YL, Pei D, et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med. 2014;371:1005–15.
Kawamata N, Pennella MA, Woo JL, Berk AJ, Koeffler HP. Dominant-negative mechanism of leukemogenic PAX5 fusions. Oncogene. 2012;31:966–77.
Fazio G, Palmi C, Rolink A, Biondi A, Cazzaniga G. PAX5/TEL acts as a transcriptional repressor causing down-modulation of CD19, enhances migration to CXCL12, and confers survival advantage in pre-BI cells. Cancer Res. 2008;68:181–9.
Fazio G, Cazzaniga V, Palmi C, Galbiati M, Giordan M, te Kronnie G, et al. PAX5/ETV6 alters the gene expression profile of precursor B cells with opposite dominant effect on endogenous PAX5. Leukemia. 2013;27:992–5.
Fortschegger K, Anderl S, Denk D, Strehl S. Functional heterogeneity of PAX5 chimeras reveals insight for leukemia development. Mol Cancer Res. 2014;12:595–606.
Schinnerl D, Fortschegger K, Kauer M, Marchante JR, Kofler R, Den Boer ML, et al. The role of the Janus-faced transcription factor PAX5-JAK2 in acute lymphoblastic leukemia. Blood. 2014. doi:10.1182/blood-2014-04-570960.
Moricke A, Reiter A, Zimmermann M, Gadner H, Stanulla M, Dordelmann M, et al. Risk-adjusted therapy of acute lymphoblastic leukemia can decrease treatment burden and improve survival: treatment results of 2169 unselected pediatric and adolescent patients enrolled in the trial ALL-BFM 95. Blood. 2008;111:4477–89.
Moricke A, Zimmermann M, Reiter A, Henze G, Schrauder A, Gadner H, et al. Long-term results of five consecutive trials in childhood acute lymphoblastic leukemia performed by the ALL-BFM study group from 1981 to 2000. Leukemia. 2010;24:265–84.
Psort II Prediction [http://www.genscript.com/psort/psort2.html]
NCBI_Protein [http://www.ncbi.nlm.nih.gov/protein/NP_036326.2]
NCBI_HomoloGene [http://www.ncbi.nlm.nih.gov/homologene?LinkName=nuccore_homologene&from_uid=302129636]
Gawin B, Niederfuhr A, Schumacher N, Hummerich H, Little PF, Gessler M. A 7.5 Mb sequence-ready PAC contig and gene expression map of human chromosome 11p13-p14.1. Genome Res. 1999;9:1074–86.
Uhlen MOP, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–50.
Strehl S, Konig M, Dworzak MN, Kalwak K, Haas OA. PAX5/ETV6 fusion defines cytogenetic entity dic(9;12)(p13;p13). Leukemia. 2003;17:1121–3.
Denk D, Nebral K, Bradtke J, Pass G, Moricke A, Attarbaschi A, et al. PAX5-AUTS2: a recurrent fusion gene in childhood B-cell precursor acute lymphoblastic leukemia. Leuk Res. 2012;36:e178–81.
Sulong S, Moorman AV, Irving JA, Strefford JC, Konn ZJ, Case MC, et al. A comprehensive analysis of the CDKN2A gene in childhood acute lymphoblastic leukemia reveals genomic deletion, copy number neutral loss of heterozygosity, and association with specific cytogenetic subgroups. Blood. 2009;113:100–7.
Zachariadis V, Gauffin F, Kuchinskaya E, Heyman M, Schoumans J, Blennow E, et al. The frequency and prognostic impact of dic(9;20)(p13.2;q11.2) in childhood B-cell precursor acute lymphoblastic leukemia: results from the NOPHO ALL-2000 trial. Leukemia. 2011;25:622–8.
Zachariadis V, Schoumans J, Barbany G, Heyman M, Forestier E, Johansson B, et al. Homozygous deletions of CDKN2A are present in all dic(9;20)(p13.2;q11.2)-positive B-cell precursor acute lymphoblastic leukaemias and may be important for leukaemic transformation. Br J Haematol. 2012;159:488–91.
Forestier E, Gauffin F, Andersen MK, Autio K, Borgstrom G, Golovleva I, et al. Clinical and cytogenetic features of pediatric dic(9;20)(p13.2;q11.2)-positive B-cell precursor acute lymphoblastic leukemias: a Nordic series of 24 cases and review of the literature. Gene Chromosome Canc. 2008;47:149–58.
Pichler H, Moricke A, Mann G, Teigler-Schlegel A, Niggli F, Nebral K, et al. Prognostic relevance of dic(9;20)(p11;q13) in childhood B-cell precursor acute lymphoblastic leukaemia treated with Berlin-Frankfurt-Munster (BFM) protocols containing an intensive induction and post-induction consolidation therapy. Br J Haematol. 2010;149:93–100.
An Q, Wright SL, Konn ZJ, Matheson E, Minto L, Moorman AV, et al. Variable breakpoints target PAX5 in patients with dicentric chromosomes: a model for the basis of unbalanced translocations in cancer. Proc Natl Acad Sci U S A. 2008;105:17050–4.
Paulsson K, Haferlach C, Fonatsch C, Hagemeijer A, Andersen MK, Slovak ML, et al. The idic(X)(q13) in myeloid malignancies: breakpoint clustering in segmental duplications and association with TET2 mutations. Hum Mol Genet. 2010;19:1507–14.
Mitelman F, Johansson B, Mertens F, (Eds.). Mitelman Database of Chromosome Aberrations and Gene Fusions in Cancer. [http://cgap.nci.nih.gov/Chromosomes/Mitelman]
Seegmiller AC, Garcia R, Huang R, Maleki A, Karandikar NJ, Chen W. Simple karyotype and bcl-6 expression predict a diagnosis of Burkitt lymphoma and better survival in IG-MYC rearranged high-grade B-cell lymphomas. Mod Pathol. 2010;23:909–20.
Mullighan CG, Su X, Zhang J, Radtke I, Phillips LA, Miller CB, et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med. 2009;360:470–80.
Moorman AV, Enshaei A, Schwab C, Wade R, Chilton L, Elliott A, et al. A novel integrated cytogenetic and genomic classification refines risk stratification in pediatric acute lymphoblastic leukemia. Blood. 2014;124:1434–44.
Abe A, Katsumi A, Kobayashi M, Okamoto A, Tokuda M, Kanie T, et al. A novel RUNX1-C11orf41 fusion gene in a case of acute myeloid leukemia with a t(11;21)(p14;q22). Cancer Genet. 2012;205:608–11.
Jones DT, Kocialkowski S, Liu L, Pearson DM, Backlund LM, Ichimura K, et al. Tandem duplication producing a novel oncogenic BRAF fusion gene defines the majority of pilocytic astrocytomas. Cancer Res. 2008;68:8673–7.
Acknowledgements
We would like to thank K. Nebral and A. Mecklenbräuker for their help with SNP data analysis. This work was supported by a grant from the Austrian Science Fund (FWF P21554-B19 to S.S.) and the St. Anna Kinderkrebsforschung e.V.
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
SA conducted experiments, analyzed the data, and drafted the manuscript; MK conducted FISH analysis; AA is responsible for the clinical data; SS supervised the study and wrote the manuscript. All authors read and approved the final manuscript.
Additional file
Additional file 1:
Summary of copy number alterations.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Anderl, S., König, M., Attarbaschi, A. et al. PAX5-KIAA1549L: a novel fusion gene in a case of pediatric B-cell precursor acute lymphoblastic leukemia. Mol Cytogenet 8, 48 (2015). https://doi.org/10.1186/s13039-015-0138-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13039-015-0138-3