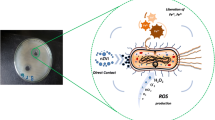
Abstract
Background
Zero-valent iron nanoparticles (ZVI NPs) have been used extensively for the remediation of contaminated soil and groundwater. Owing to their large active surface area, they serve as strong and effective reductants. However, the ecotoxicity and bioavailability of ZVI NPs in diverse ecological media have not been evaluated in detail and most studies have focused on non-nano ZVI or Fe0. In addition, the antimicrobial properties of ZVI NPs have rarely been investigated, and the underlying mechanism of their toxicity remains unknown.
Results
In the present study, we demonstrate that ZVI NPs exhibited significant toxicity at 1000 ppm against two distinct gram-positive bacterial strains (Bacillus subtilis 3610 and Bacillus thuringiensis 407) but not against two gram-negative strains (Escherichia coli K12 and ATCC11634). Specifically, ZVI NPs caused at least a 4-log and 1-log reductions in cell numbers, respectively, in the two Bacillus strains, whereas no change was detected in the two E. coli strains. X-ray photoelectron spectroscopy, X-ray absorption near-edge, and extended X-ray absorption fine structure spectra confirmed that Bacillus cells exposed to ZVI NPs contained mostly Fe2O3 with some detectable FeS. This finding indicated that Fe0 nanoparticles penetrated the bacterial cells, where they were subsequently oxidized to Fe2O3 and FeS. RedoxSensor analysis and propidium iodide (PI) staining showed decreased reductase activity and increased PI in both Bacillus strains treated with a high (1000 ppm) concentration of ZVI NPs.
Conclusion
Taken together, these data show that the toxicity of ZVI NPs was derived from their oxidative properties, which may increase the levels of reactive oxygen species and lead to cell death.
Similar content being viewed by others
Background
Zero-valent iron nanoparticles (ZVI NPs) have been extensively used to remediate contaminated soil and groundwater because of their ability to target chlorinated organic compounds (e.g., polychlorinated biphenyl, trichloroethylene, perchloroethylene, pesticides, solvents), inorganic anions (e.g., perchlorate), and heavy metals [1,2,3]. ZVI NPs have a large active surface area, can serve as strong and effective reductants, and show an elevated capacity to absorb abundant heavy metals and contaminants [4]. There are many types of ZVI NPs, including uncapped, surface-modified, and bimetallic ZVI NPs, which can be used for the dechlorination of chlorinated solvents, in dense non-aqueous phase liquid source zones, and for odor control in biosolid treatments [5,6,7,8]. Other metal nanoparticles, such as silver, gold, and copper nanoparticles [9,10,11] have been reported to negatively affect bacterial growth and to kill bacteria in the soil [12, 13], through mechanisms that have been only partially characterized. At present, the toxicity of ZVI NPs towards bacteria and the underlying mechanism remain unclear.
The ecotoxicity and bioavailability of ZVI NPs in diverse ecological media have not been evaluated in detail, and the studies conducted to date have mostly focused on non-nano ZVI or Fe0. Although ZVI NPs themselves are nontoxic, they can oxidize Fe0 to Fe2+ and then to Fe3+, and these chemical forms are readily found in the habitats of soil microorganisms. In addition, application of ZVI NPs under oxidative conditions can considerably increase the local concentrations of Fe2+ and Fe3+. Consequently, oxidation of ZVI NPs leads to the production of reactive oxygen species (ROS), resulting in the generation of hydroxyl radicals (OH−) from superoxide (O2 −) and hydrogen peroxide (H2O2) in microbial cells. These radicals enhance oxidative stress and cause cell membrane damage, leading to the outflow of intracellular contents and, ultimately, cell death [9, 14].
To date, there have been few reports on the toxicity of ZVI NPs. Some studies have shown the ability of ZVI NPs to inhibit the growth of Escherichia coli by disrupting its cell membrane [15,16,17], whereas other studies have shown that ZVI NPs smaller than 50 nm were not effective against the soil gram-negative bacterium Pseudomonas stutzeri, even when treated at 10,000 ppm for 48 h [18]. Diao and Yao [9] found that 20–30-nm ZVI NPs caused a 100% reduction (1000–10,000 ppm) in Pseudomonas fluorescens and 80–100% reduction (100–10,000 ppm) in Bacillus subtilis but had no effect (100–10,000 ppm) on the growth of Aspergillus versicolor.
There have been very few reports on the influence of ZVI NPs on complex microbial ecosystems [19,20,21,22,23,24]. It is therefore difficult to predict the effects of NPs on an ecological system, given that their behavior and environmental fate are poorly understood [13]. Considering the high daily fluctuations in environmental soil parameters, this represents an additional challenge to realistically simulate the complex soil environment in an experimental exposure scenario. Indeed, the mobility and toxicity of metal particles to soil microorganisms can be influenced by several natural soil parameters, such as pH, calcium concentration, organic matter content, C-to-N ratio, and cation exchange capacity [25, 26]. However, ZVI NPs can also have an inhibitory effect on biological dechlorination in the presence of 1 g/L iron nanoparticles, resulting in toxicity to indigenous bacteria that hinders their participation in the remediation process [9, 27]. Xiu et al. [27] found that ZVI NPs initially inhibited the growth of bacteria dechlorinating trichloroethylene, but their dechlorination activity and ethane production recovered after a lag period [27]. Given that ZVI NPs are highly redox-active metals [28], their environmental impact and safety when applied to the soil should be investigated in more detail [12, 29].
In the present study, we examined the antimicrobial activity of different concentrations of ZVI NPs against two common soil gram-positive bacteria, Bacillus subtilis 3610 and B. thuringiensis 407, as well as two common soil gram-negative bacteria, Escherichia coli K12 and ATCC11634, grown in Mueller Hinton broth (MHB). The potential mechanisms underlying toxicity were examined by X-ray absorption spectroscopy, which is an excellent tool to determine valence and local structure.
Methods
ZVI NP preparation
Ten grams of FeSO4·7H2O powder was dissolved in 70 mL deionized water with stirring at 220–260 rpm and then mixed with 30 mL ethanol. Next, 2.0 M NaOH aqueous solution was added dropwise until a pH value of 6.8 ± 0.1 was achieved. Next, 1.8 g of NaBH4 was dissolved in 5.0 mL deionized water and dropped in the solution with mild stirring for 30 min. A magnet was applied to separate the nano-iron particles thus generated from the solution. The filtered ZVI NPs were washed using deionized water and ethanol three times each. Finally, ZVI NPs were dried at room temperature using flowing N2 for 12 h.
X-ray absorption near-edge structure (XANES) and extended X-ray absorption fine structure (EXAFS) analyses
Overnight cultures containing approximately 1 × 109 colony-forming units (CFU)/mL of E. coli K12, E. coli ATCC11634, B. subtilis 3610, and B. thuringiensis 407 were diluted 100-fold into 250 mL of MHB in four 500-mL Pyrex flasks. ZVI NPs were added to a concentration of 1000 ppm. Cells were grown for 3 h at 37 °C and 175 rpm in a rotary shaker, after which they were centrifuged at 5000 rpm for 15 min, the supernatant was discarded, and cells were washed with double-distilled H2O. Finally, samples were freeze-dried into powder. XANES/EXAFS spectra were collected on the Wiggler beam line 17C1 at the National Synchrotron Radiation and Research Center (NSRRC) of Taiwan. The electron storage ring was operated at an energy of 1.5 GeV and a ring current of 100–200 mA. Data were collected in transmission mode by a Lytle detector in the region of the Fe K-edge at 25 °C. Data were subsequently normalized using Athena (vi) software, with the linear pre-edge and polynomial post-edge backgrounds subtracted from the raw ln(I t /I 0) data (where I t is the light intensity after it passes through the sample and I 0 is the initial light intensity), and then analyzed using Artemis (vi) software with FFEF-8. The spectra were first energy-calibrated by simultaneously recording the transmission spectra of the Fe foil with Athena (vi), where the energy of the first inflection point for the reference sample absorption edge was 7112 eV. After calibration, samples were background-corrected (using a linear pre-edge region and a polynomial for the post-edge region) and normalized. EXAFS energy spectra were then converted into wavevector K-space form. Data directly reflected the average local environment around the absorbing atoms. Spectra were analyzed using the IFEFFIT [30, 31] software package. The theoretical paths for the Fe–Fe and Fe–O species were generated using the FEFF-8 program based on the crystallographic data of the individual species and were used to fit the first coordination shell of the experimental data. The coordination number, interatomic distance, Debye–Waller factor, and inner potential corrections were used as variables for the fitting procedures.
ZVI NP characterization
The X-ray diffraction (XRD) patterns of the synthesized ZVI NP samples were recorded at a scan rate of 4° (2θ)/min using monochromatic Cu Kα radiation (MXP18; MAC Science, Japan) at 30 kV and 20 mA. The recorded specific peak intensity and 2θ values were further identified using a database system (JCPDS). The morphology, microstructure, and particle size of the ZVI NPs were investigated by field-emission scanning electron microscopy (FE-SEM; S-4700 Type II; Hitachi, Japan) and high-resolution transmission electron microscopy (HR-TEM; H-7500; Hitachi, Japan).
Assessment of the antibacterial effects of ZVI NPs, Fe2O3, and oxidized Fe NPs
Escherichia coli K12, E. coli ATCC11634, B. subtilis 3610, and B. thuringiensis 407 strains were maintained in MHB on 1.5% Bacto agar plates at 37 °C. Overnight cultures of approximately 5 × 108 CFU/mL were added in 50 mL MHB in 250-mL Pyrex flasks at pH 7. ZVI NPs were added to final concentrations of 0, 100, 500, and 1000 ppm. Cells were grown for 3 h at 37 °C and 175 rpm in a rotary shaker. All cultures were serially diluted, plated on Luria–Bertani (LB) agar plates, incubated overnight at 37 °C, and then subjected to a colony count. All experiments were performed in duplicate, and each value represents the mean of three technical replicates. Statistical significance was determined using one-way analysis of variance (ANOVA).
RedoxSensor measurements
Reductase activities of B. subtilis 3610 and B. thuringiensis 407 were determined using a BacLight RedoxSensor Green Vitality Kit (ThermoFisher, Waltham, MA, USA). Overnight cultures of approximately 1 × 107 CFU/mL in 50 mL MHB were supplemented with the indicated concentrations of ZVI NPs for 3 h at 175 rpm and 37 °C. Cells were washed with 1 × phosphate-buffered saline (PBS) twice, diluted 10-fold with the same buffer, and eventually mixed well with 1 μL of RedoxSensor Green reagent. Additionally, 1 μL of propidium iodide (PI) was added to the mixture, which was incubated in the dark at room temperature for 5 min before assessing cell membrane integrity. Stained cells (1 mL) in PBS were assayed with a FACSCalibur Flow Cytometer (BD Biosciences, San Jose, CA, USA). In addition, fluorescence filters and detectors were all standardized with green fluorescence collected in the FL1 channel (530 ± 15 nm) and red fluorescence collected in the FL3 channel (> 650 nm). Data were analyzed using FACSCalibur Flow Cytometer software. All parameters were collected as logarithmic signals. All measurements were performed in two separate experiments.
X-ray photoelectron spectroscopy (XPS) analyses
The chemical composition on the surface of the as-synthesized ZVI NPs was determined by XPS (Physical Electronic ESCA PHI 1600; Chanhassen, MN, USA) at the excitation energy of Al Kα (1486.6 eV). The C 1s (284.5 eV) peak was used as the calibration standard for the wide-region spectra of these samples with different Fe valences [e.g. Fe(0), Fe(II), Fe(III)]. XPS signals of the above species were recorded with a cylindrical mirror analyzer (CMA). The fractions of these samples with different Fe valences were calculated by their integrated peak areas. All measurements were performed in two separate experiments.
Results
ZVI NPs inhibited growth
ZVI NPs vary broadly in terms of size and morphology [32,33,34]. Thus, to compare the present results with those of previous studies, we first characterized the size and structure of the ZVI NPs synthesized using FE-SEM. As shown in Fig. 1a, b, ZVI NPs were about 4.8 nm in diameter on average. XRD patterns showed a main characteristic diffraction peak at 2θ = 44.7° (Fig. 1c) based on the diffraction patterns for Fe0 nanoparticles (JCPDS Card Number 06-0696, [35]). The overall XRD pattern was comparable with previously reported spectra.
Next, we examined the antimicrobial activity of ZVI NPs against two gram-positive soil bacteria (B. subtilis 3610 and B. thuringiensis 407) and two gram-negative soil bacteria (E. coli K12 and ATCC11634). ZVI NPs did not significantly retard the growth of either E. coli strain, but inhibited both Bacillus strains (Fig. 2a). Specifically, B. subtilis 3610 exhibited a 4-log reduction, and B. thuringiensis 407 showed a 1-log reduction in the presence of 1000 ppm ZVI NPs (Fig. 2a), indicating that the latter was relatively more resistant. Collectively, these data suggested that ZVI NPs were more toxic to gram-positive than gram-negative bacteria. In addition, we next aimed to differentiate whether ZVI NP oxide entered cells and caused ROS generation, leading to cell death, or whether oxide ZVI NPs, such as Fe2O3, caused cell death. Therefore, ZVI NPs were shaken in MHB medium under aerobic conditions for 3 h, and we then examined oxidation forms. XPS and XANES analyses showed that most ZVI NPs became Fe2O3. We further used oxide ZVI NPs and Fe2O3 in antimicrobial assays and found that even 1000 ppm oxide ZVI NPs and Fe2O3 were unable to inhibit cell growth or death for all four strains (Fig. 2b, c). This suggested that ZVI NPs killed bacterial cells when the ZVI NPs entered cells and caused oxidation and ROS generation, thereby leading to cell death.
Antibacterial effects of different ZVI NP concentrations against E. coli and Bacillus strains. a B. subtilis 3610 (Bs 3610), B. thuringiensis 407 (Bt 407), E. coli K12 (Ec K12), and E. coli ATCC11634 (Ec ATCC11634) grown in MHB medium were treated with different concentrations of ZVI NPs at 37 °C with shaking at 175 rpm for 3 h; antibacterial activity was evaluated by plating and counting cells. B. subtilis 3610, B. thuringiensis 407, E. coli K12, and E. coli ATCC11634 grown in MHB medium were treated with different concentrations of b Fe2O3 c Oxidized Fe(ZVI) NPs at 37 °C with shaking at 175 rpm for 3 h; antibacterial activity was evaluated as above. Data are expressed as means ± standard deviations of two separate experiments, with three replicates. **p < 0.01 as determined by ANOVA relative to the control sample at different pH values
ZVI NPs altered the redox status in B. subtilis and B. thuringiensis
ZVI NPs may cause an increase in ROS; therefore, to examine the potential mechanisms of the observed growth inhibition [28], we evaluated whether these nanoparticles could affect reductase activity in the two Bacillus strains. Briefly, overnight bacterial cell cultures (~ 1 × 107 CFU/mL) were treated with 0–1000 ppm ZVI NPs for 3 h and then stained with RedoxSensor Green, consisting of a green fluorescent dye to assess oxidoreductase activity and a red dye to assess membrane integrity. Accordingly, ZVI NPs reduced the percentage of cells (M1 Gate %) showing reductase activity in a concentration-dependent manner (Table 1, Fig. 3a) and dramatically increased the percentage of PI-positive cells (M1 Gate %; Fig. 3b). Thus, nanoparticles severely compromised the integrity of the cell membranes after 3 h, suggesting that, compared with the control, many cells were dead at 1000 ppm (Table 1, Fig. 3b).
Analysis of RedoxSensor and propidium iodide (PI) staining intensity in B. subtilis 3610 (Bs3610) and B. thuringiensis 407 (Bt407). The two Bacillus strains were grown in MHB supplemented with ZVI NPs at concentrations of 0, 100, 500, and 1000 ppm for 3 h, and the cells were then treated with a RedoxSensor Green (green) or b PI (red). The X-axis shows the RedoxSensor or PI fluorescence intensity in arbitrary units (au), and the Y-axis indicates the cell counts as measured by flow cytometry. PBS-only and unstained cells were used as controls. Data are representative of two separate experiments. RedoxSensor fluorescence is presented in a false green color, and PI staining is presented in a false red color for clearer visualization
Oxidation states and fine structural parameters of ZVI NPs in B. subtilis 3610 and B. thuringiensis 407
To resolve whether the observed toxicity was derived from the ZVI NPs themselves or the consequent iron oxidation and production of intracellular ROS, we analyzed the Fe oxidation states of ZVI in the two Bacillus strains and two E. coli strains using XPS [36]. The findings are presented in Table 2 as the integrated area percentages of Fe(0), Fe(II), and Fe(III). The iron oxidation states on the surfaces of these samples were analyzed using XPS spectra, as illustrated in Table 2. In particular, we found that the ZVI NPs were oxidized to Fe(III) (60.8%) or Fe(II) (39.2%) after 3-h oxidation treatment in MHB medium. Since the ZVI NPs came in contact with dissolved oxygen from the atmosphere, it would be oxidized to iron oxides, such as FeO, Fe2O3, and Fe3O4. Thus, this phenomenon implied the high reduction ability of ZVI NPs in MHB medium for killing the bacteria. The iron species in the B. subtilis 3610 and B. thuringiensis 407 were identified as Fe(0) (17.6 and 11.9%), Fe(II) (27.0 and 26.9%), and Fe(III) (55.4 and 61.2%), respectively. The existence of Fe(0) was attributed to rapid absorption or diffusion of ZVI NPs into B. subtilis 3610 and B. thuringiensis 407, reducing the oxidization of ZVI NPs. In contrast, ZVI NPs in solution could not easily be absorbed or diffused through the outer bilayer cell membranes of E. coli K12 and E. coli ATCC11634, thus suppressing cytotoxicity.
Furthermore, the iron oxidation states in the internals of all Bacillus and E. coli cells were investigated with XANES spectra [37] in Fig. 4a–e. In comparison to iron standards (Fe, FeO, Fe2O3, and Fe3O4), it could be seen that the iron valence in all Bacillus and E. coli cells belonged to Fe(III). These results suggested that Fe0 NPs did enter the cells and became oxidized into FeO, Fe2O3, and Fe3O4. The XANES spectra of two Bacillus and two E. coli strains overlapped with the spectra of Fe2O3 and Fe3O4 from 7100 to 7200 eV, particularly at the absorption peak of the iron species (7135 eV). To compare the oxidation states for the two strains and the Fe2O3 and Fe3O4 standards, the absorption peaks from 7125 to 7160 eV were enlarged, as shown in Fig. 4a–e. The absorption peaks of both strains at 7135 eV were similar to those of Fe2O3, indicating that iron had valence Fe(III) in both cases. However, it is difficult to compare the valence of iron species between the two E. coli strains (Fig. 4d, e).
Normalized K-edge XANES spectra of ZVI NPs standards, Bacillus cells, and E. coli cells treated with 1000 ppm ZVI NPs. K-edge-normalized XANES spectra of a B. subtilis 3610 (Bs 3610), b B. thuringiensis 407 (Bt 407), c oxidized Fe(ZVI) NPs, d E. coli K12 (Ec K12), and e E. coli ATCC11634 (Ec ATCC11634). Data are representative of two separate experiments
To further characterize and confirm Fe2O3 in the two Bacillus strains, two E. coli strains, and oxidized ZVI NPs, EXAFS spectra were obtained. As shown in Fig. 5a–e, the Fe–O bond distance of the first shell in both Bacillus strains was consistent with that of the Fe2O3 standard. Therefore, the atomic neighbors, bond distance, and coordination number of Fe2O3 in the two Bacillus strains, two E. coli strains, and oxidized ZVI NPs could be analyzed according to the fine structural parameters of Fe2O3. A similar overlap with the Fe2O3 standard was observed also for k3-weighted and least squares-fitted Fe2O3 K-edge fine structure inverse Fourier-transform oscillation spectra of both strains (Fig. 6a–e). Detailed structural parameters of Fe2O3 in two Bacillus strains, two E. coli strains, and oxidized ZVI NPs are given in Table 3. R-factor values were much smaller than 0.02, demonstrating the great reliability of EXAFS analysis. Generally, the coordination number (CN) of iron atoms is larger in oxides than that in metals. It is ascribed to the occupation of oxygen atoms near central iron atom in crystal structure. In particular, the coordination number for B. subtilis 3610 was larger than that for B. thuringiensis 407 and E. coli strains, indicating more neighboring oxygen atoms around the central Fe atoms in B. subtilis 3610. Accordingly, these extra oxygen atoms occupy the limited crystal structure space, which explains the shorter Fe–O bond distance than that detected in B. thuringiensis 407 and E. coli strains. Overall, the results of XANES and EXAFS analyses suggested that ZVI NPs were likely oxidized when in contact with the oxygen present inside bacteria, which then caused their death.
Discussion
In this study, we found that 1000 ppm ZVI NPs was lethal to the two Bacillus strains but not to the two E. coli strains tested. At 1000 ppm, B. thuringiensis 407 was more resistant than B. subtilis 3610 to ZVI NPs, whereas no significant decrease was observed for either when treated with 500 ppm ZVI NPs. These results suggested that gram-negative strains may be more resistant to ZVI NPs than gram-positive strains.
Lee et al. [16] reported that 9 ppm ZVI NPs of 10–80 nm (average diameter ~ 35 nm) were toxic to gram-negative E. coli ATCC8739, resulting in 2.3-log inactivation per mg/L/h of cells grown in 2 mM carbonate buffer at pH 8.0 in the absence of oxygen. However, under aerobic conditions, 90 ppm ZVI NPs caused around 0.029-log inactivation per mg/L/h of cells. This finding suggested that ZVI NPs might exert significant antimicrobial effect only under anaerobic conditions. Notably, Lee et al. [16] used an atypical antimicrobial assay with an artificial carbonate buffer solution at a relatively more alkaline pH (pH 8), which was assessed under anaerobic conditions. This setting is not ideal for determining the practical minimum bactericidal concentration because E. coli does not grow very well in the absence of oxygen and at pH 8. In nature, E. coli cells live in aerobic environments with a pH close to 7. Thus, this method is likely too artificial to accurately measure cell death. Instead, in this study, we used the minimum bactericidal concentration method, a more common antimicrobial assay. Viability was determined by plating the cells and counting cell numbers. Even with 1000 ppm ZVI NPs at pH 7 and pH 8 (data not shown), the viability of both E. coli strains was comparable to that of untreated controls, whereas the viability of both Bacillus strains decreased significantly when treated with either 500 or 1000 ppm ZVI NPs at pH 7.
Diao and Yao [9] found that treatment with 100–1000 ppm of 20–30-nm ZVI NPs reduced B. subtilis viability by 80–100%, representing a 2-log reduction. In contrast, in the present study we found a 4-log reduction in viability with 1000 ppm ZVI NPs of a smaller size. This discrepancy suggested that size may influence the ZVI NP antimicrobial effect, with different species and strains displaying variations in the extent of resistance or susceptibility to ZVI NPs. Despite the clear difference in resistance between gram-positive and gram-negative strains reported here, gram-negative P. stutzeri is resistant to ZVI NPs, whereas E. coli and P. fluorescens are more sensitive [18], making it difficult to conclude that gram-negative strains are more sensitive to ZVI NPs.
To elucidate the mechanism through which ZVI NPs decrease bacterial cell growth, we stained Bacillus cells with the fluorescent dyes RedoxSensor and PI to test whether the nanoparticles decreased reductase activity and disrupted bacterial cell membranes. The observed concentration-dependent increase in membrane damage suggested that the nanoparticles increased membrane permeability and possibly penetrated cells to cause protein toxicity. Moreover, the observed decrease in reductase activity after exposure to ZVI NPs suggested that increased levels of ROS contributed to loss of membrane integrity and cell viability.
Therefore, the high concentrations of ROS generated by ZVI NPs may have the most deleterious effects on bacterial cells [36, 38,39,40]. ROS damage iron-sulfur groups in the cofactors of many enzymes, leading to the production of even more ROS via the Fenton reaction and resulting in cell injury and death. Importantly, we have now clearly demonstrated that ZVI NP-induced toxicity is related to an increase in oxidation and ROS. Upon internalization, ZVI NPs may be oxidized to Fe2O3, which could further promote ROS production to alter the redox status and kill the bacterial cells [41]. This process may explain the observed decrease in reductase activity and the increase in PI staining in both Bacillus strains.
XANES/EXAFS measurements showed that Fe2O3 was present in high amounts in bacterial cells incubated with ZVI NPs. In addition, all cells incubated with pre-oxidized ZVI NPs that became Fe2O3 were nontoxic to all strains, suggesting that ZVI NPs entered cells and were oxidized to Fe2O3, resulting in cell death, but were not oxidized outside cells. This was further confirmed with the PI and RedoxSensor assays. Overall, our results suggested a toxic mechanism through which ZVI NPs enter Bacillus cells and induce a ROS response, resulting in decreased RedoxSensor activity, with subsequent oxidation to Fe2O3.
However, E. coli strains were more resistant to ZVI NPs or the ROS response. On the basis of XPS, XANES, and EXAFS analyses, ZVI NPs were completely oxidized to FeO (Fe(II)) in MHB medium within a short time and then continuously oxidized, forming a relatively thick outer layer of Fe2O3 (Fe(III)). Moreover, the ZVI NPs were absorbed through monolayer cell membranes of B. subtilis 3610 and B. thuringiensis 407 and then became oxidized to FeO and Fe2O3 within the cells. Some ZVI NPs were accumulated inside of B. subtilis 3610 and B. thuringiensis 407 and could be easily detected by XPS. During the duration of ZVI NP oxidation, electrons were generated, exciting the chemical species inside bacteria into free radicals. Subsequently, B. subtilis 3610 and B. thuringiensis 407 were killed by these internal free radicals. Apart from the generation of free radicals, the consumption of oxygen within cells may be another reason for the death of B. subtilis 3610 and B. thuringiensis 407. For E. coli K12 and E. coli ATCC11634, ZVI NPs could not easily attach to the cell surface. Only a few ZVI NPs were oxidized near the surface of E. coli K12 and E. coli ATCC11634 and then coincidentally entered the cells via tiny cracks in the bilayer cell membrane.
This might explain the results observed in Bacillus cultures treated with ≥ 500 ppm ZVI NPs, whereby RedoxSensor activity was significantly decreased but membrane permeability was significantly increased. To the best of our knowledge, this is the first study to directly analyze ZVI NPs internalized in bacterial cells using XANES and EXAFS and to demonstrate that ZVI NP oxidation was the primary source of their toxicity [30, 31, 42]. ZVI NPs have been used for different applications in soil remediation [43,44,45]. The findings of this study provide new molecular insights into the effects of ZVI NP toxicity on gram-positive and gram-negative bacteria for application in ecotoxicological tests.
Conclusion
To the best of our knowledge, this is the first study demonstrating the toxicity of ZVI NPs (at 1000 ppm) against gram-positive B. subtilis and B. thuringiensis but not gram-negative E. coli strains in the soil. In addition, XANES/EXAFS data showed that Fe2O3 was present in high amounts in the Bacillus cells cultured with ZVI NPs. Our findings suggested that ZVI NPs enter Bacillus cells and induce a ROS response, resulting in decreased RedoxSensor activity, with subsequent oxidation to Fe2O3. ZVI NPs have been used for different applications in soil remediation [43,44,45]; however, few studies have looked at the persisting challenges of such use and evaluated the biological interactions of NPs. The present findings may provide a molecular approach to elucidate the effects of ZVI NP toxicity on gram-positive and gram-negative bacteria for applications in ecotoxicological tests.
Abbreviations
- ZVI NPs:
-
zero-valent iron nanoparticles
- ROS:
-
reactive oxygen species
- MHB:
-
Mueller–Hinton broth
- FE-SEM:
-
field-emission scanning electron microscopy
- XRD:
-
X-ray diffraction
- PI:
-
propidium iodide
- XPS:
-
X-ray photoelectron spectroscopy
- XANES:
-
X-ray adsorption near-edge spectra
- EXAFS:
-
extended X-ray adsorption fine structure
- HR-TEM:
-
high-resolution transmission electron microscopy
- LB:
-
Luria–Bertani
- ANOVA:
-
analysis of variance
- CMA:
-
cylindrical mirror analyzer
- PBS:
-
phosphate-buffered saline
References
Cao J, Elliott D, Zhang W-x. Perchlorate reduction by nanoscale iron particles. J Nanopart Res. 2005;7:499–506.
Grieger KD, Fjordboge A, Hartmann NB, Eriksson E, Bjerg PL, Baun A. Environmental benefits and risks of zero-valent iron nanoparticles (nZVI) for in situ remediation: risk mitigation or trade-off? J Contam Hydrol. 2010;118:165–83.
Ponder F Jr, Li F, Jordan D, Berry EC. Assessing the impact of Diplocardia ornata on physical and chemical properties of compacted forest soil in microcosms. Biol Fert Soils. 2000;32:166–72.
Johnson RL, Johnson GOB, Nurmi JT, Tratnyek PG. Natural organic matter enhanced mobility of nano zerovalent iron. Environ Sci Technol. 2009;43:5455–60.
Bezbaruah AN, Krajangpan S, Chisholm BJ, Khan E, Bermudez JJ. Entrapment of iron nanoparticles in calcium alginate beads for groundwater remediation applications. J Hazard Mater. 2009;166:1339–43.
Fu F, Dionysiou DD, Liu H. The use of zero-valent iron for groundwater remediation and wastewater treatment: a review. J Hazard Mater. 2014;267:194–205.
Gong X, Li W, Wang K, Hu J. Study of the adsorption of Cr(VI) by tannic acid immobilised powdered activated carbon from micro-polluted water in the presence of dissolved humic acid. Bioresour Technol. 2013;141:145–51.
Xie T, Reddy KR, Wang C, Yargicoglu E, Spokas K. Characteristics and applications of biochar for environmental remediation: a review. Crit Rev Environ Sci Tech. 2015;45:939–69.
Diao M, Yao M. Use of zero-valent iron nanoparticles in inactivating microbes. Water Res. 2009;43:5243–51.
Schrand AM, Rahman MF, Hussain SM, Schlager JJ, Smith DA, Syed AF. Metal-based nanoparticles and their toxicity assessment. Wires Nanomed Nanobi. 2010;2:544–68.
Shankar S, Rhim J-W. Effect of copper salts and reducing agents on characteristics and antimicrobial activity of copper nanoparticles. Mater Lett. 2014;132:307–11.
Farre M, Gajda-Schrantz K, Kantiani L, Barcelo D. Ecotoxicity and analysis of nanomaterials in the aquatic environment. Anal Bioanal Chem. 2009;393:81–95.
Klaine SJ, Alvarez PJ, Batley GE, Fernandes TF, Handy RD, Lyon DY, Mahendra S, McLaughlin MJ, Lead JR. Nanomaterials in the environment: behavior, fate, bioavailability, and effects. Environ Toxicol Chem. 2008;27:1825–51.
Barnes RJ, van der Gast CJ, Riba O, Lehtovirta LE, Prosser JI, Dobson PJ, Thompson IP. The impact of zero-valent iron nanoparticles on a river water bacterial community. J Hazard Mater. 2010;184:73–80.
Auffan M, Achouak W, Rose J, Roncato M-A, Chanéac C, Waite DT, Masion A, Woicik JC, Wiesner MR, Bottero J-Y. Relation between the redox state of iron-based nanoparticles and their cytotoxicity toward Escherichia coli. Environ Sci Technol. 2008;42:6730–5.
Lee C, Kim JY, Lee WI, Nelson KL, Yoon J, Sedlak DL. Bactericidal effect of zero-valent iron nanoparticles on Escherichia coli. Environ Sci Technol. 2008;42:4927–33.
Li Z, Greden K, Alvarez PJ, Gregory KB, Lowry GV. Adsorbed polymer and NOM limits adhesion and toxicity of nano scale zerovalent iron to E. coli. Environ Sci Technol. 2010;44:3462–7.
Sacca ML, Fajardo C, Martinez-Gomariz M, Costa G, Nande M, Martin M. Molecular stress responses to nano-sized zero-valent iron (nZVI) particles in the soil bacterium Pseudomonas stutzeri. PLoS ONE. 2014;9:e89677.
Cullen LG, Tilston EL, Mitchell GR, Collins CD, Shaw LJ. Assessing the impact of nano- and micro-scale zerovalent iron particles on soil microbial activities: particle reactivity interferes with assay conditions and interpretation of genuine microbial effects. Chemosphere. 2011;82:1675–82.
Fajardo C, Ortiz LT, Rodriguez-Membibre ML, Nande M, Lobo MC, Martin M. Assessing the impact of zero-valent iron (ZVI) nanotechnology on soil microbial structure and functionality: a molecular approach. Chemosphere. 2012;86:802–8.
Keenan CR, Goth-Goldstein R, Lucas D, Sedlak DL. Oxidative stress induced by zero-valent iron nanoparticles and Fe(II) in human bronchial epithelial cells. Environ Sci Technol. 2009;43:4555–60.
Nemecek J, Lhotsky O, Cajthaml T. Nanoscale zero-valent iron application for in situ reduction of hexavalent chromium and its effects on indigenous microorganism populations. Sci Total Environ. 2014;485–486:739–47.
Tilston EL, Collins CD, Mitchell GR, Princivalle J, Shaw LJ. Nanoscale zerovalent iron alters soil bacterial community structure and inhibits chloroaromatic biodegradation potential in Aroclor 1242-contaminated soil. Environ Pollut. 2013;173:38–46.
Yang Y, Guo J, Hu Z. Impact of nano zero valent iron (NZVI) on methanogenic activity and population dynamics in anaerobic digestion. Water Res. 2013;47:6790–800.
Beyer WN, Cromartie EJ. A survey of Pb, Cu, Zn, Cd, Cr, As, and Se in earthworms and soil from diverse sites. Environ Monit Assess. 1987;8:27–36.
Kızılkaya R. The role of different organic wastes on zinc bioaccumulation by earthworm Lumbricus terrestris L. (Oligochaeta) in successive Zn added soil. Ecol Eng. 2005;25:322–31.
Xiu ZM, Jin ZH, Li TL, Mahendra S, Lowry GV, Alvarez PJ. Effects of nano-scale zero-valent iron particles on a mixed culture dechlorinating trichloroethylene. Bioresour Technol. 2010;101:1141–6.
Kumar N, Omoregie EO, Rose J, Masion A, Lloyd JR, Diels L, Bastiaens L. Inhibition of sulfate reducing bacteria in aquifer sediment by iron nanoparticles. Water Res. 2014;51:64–72.
Jang MH, Lim M, Hwang YS. Potential environmental implications of nanoscale zero-valent iron particles for environmental remediation. Environ Health Toxicol. 2014;29:e2014022.
Nesvizhskii A, Rehr J. L-edge XANES of 3d-transition metals. J Synchrotron Radiat. 1999;6:315–6.
Ravel B, Newville M. ATHENA and ARTEMIS: interactive graphical data analysis using IFEFFIT. Phys Scripta. 2005;T1005:1007.
Behera SS, Patra JK, Pramanik K, Panda N, Thatoi H. Characterization and evaluation of antibacterial activities of chemically synthesized iron oxide nanoparticles. World J Nano Sci Eng. 2012;02:196–200.
Ronavari A, Balazs M, Tolmacsov P, Molnar C, Kiss I, Kukovecz A, Konya Z. Impact of the morphology and reactivity of nanoscale zero-valent iron (NZVI) on dechlorinating bacteria. Water Res. 2016;95:165–73.
Petala E, Baikousi M, Karakassides MA, Zoppellaro G, Filip J, Tuček J, Vasilopoulos KC, Pechoušek J, Zbořil R. Synthesis, physical properties and application of the zero-valent iron/titanium dioxide heterocomposite having high activity for the sustainable photocatalytic removal of hexavalent chromium in water. Phys Chem Chem Phys. 2016;18:10637–46.
Ji J, Yan X, Qian G, Peng C, Duan X, Zhou X. Morphology and location manipulation of Fe nanoparticles on carbon nanofibers as catalysts for ammonia decomposition to generate hydrogen. Int J Hydrogen Energ. 2017;42:17466–75.
Lv Y, Niu Z, Chen Y, Hu Y. Bacterial effects and interfacial inactivation mechanism of nZVI/Pd on Pseudomonas putida strain. Water Res. 2017;115:297–308.
Li L, Hu J, Shi X, Fan M, Luo J, Wei X. Nanoscale zero-valent metals: a review of synthesis, characterization, and applications to environmental remediation. Environ Sci Pollut Res Int. 2016;23:17880–900.
Padrova K, Matatkova O, Sikova M, Fuzik T, Masak J, Cejkova A, Jirku V. Mitigation of Fe(0) nanoparticles toxicity to Trichosporon cutaneum by humic substances. N Biotechnol. 2016;33:144–52.
Ortega-Calvo JJ, Jimenez-Sanchez C, Pratarolo P, Pullin H, Scott TB, Thompson IP. Tactic response of bacteria to zero-valent iron nanoparticles. Environ Pollut. 2016;213:438–45.
Barzan E, Mehrabian S, Irian S. Antimicrobial and genotoxicity effects of zero-valent iron nanoparticles. Jundishapur J Microbiol. 2014;7:e10054.
Cheng R, Li G, Shi L, Xue X, Kang M, Zheng X. The mechanism for bacteriophage f2 removal by nanoscale zero-valent iron. Water Res. 2016;105:429–35.
Lytle FW. The EXAFS family tree: a personal history of the development of extended X-ray absorption fine structure. J Synchrotron Radiat. 1999;6:123–34.
Yang YF, Cheng YH, Liao CM. In situ remediation-released zero-valent iron nanoparticles impair soil ecosystems health: a C. elegans biomarker-based risk assessment. J Hazard Mater. 2016;317:210–20.
Pawlett M, Ritz K, Dorey RA, Rocks S, Ramsden J, Harris JA. The impact of zero-valent iron nanoparticles upon soil microbial communities is context dependent. Environ Sci Pollut Res Int. 2013;20:1041–9.
Oleszczuk P, Koltowski M. Effect of co-application of nano-zero valent iron and biochar on the total and freely dissolved polycyclic aromatic hydrocarbons removal and toxicity of contaminated soils. Chemosphere. 2017;168:1467–76.
Authors’ contributions
Designed and conceived the experiments: Y-HH. Performed the experiments: P-HT, Y-HH, W-JK, and C-LC. Analyzed the data: Y-HH, K-SL, and W-JK. Contributed reagents/materials/analysis tools: Y-HH and K-SL. Wrote the paper: Y-HH. All authors read and approved the final manuscript.
Acknowledgements
The authors appreciate the technical assistance of the Microscope Core Laboratory at Chang Gung Memorial Hospital and the technical support regarding XANES/EXAFS measurements at the National Synchrontron Radiation Research Center of Taiwan.
Competing interests
The authors declare that they have no competing interests.
Availability of data and materials
All data generated or analyzed during this study are included in the published article.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Funding
This work was supported by Grant FEMH-YZU-2017-007 from Yuan Ze University, Taiwan, to Yi-Huang Hsueh. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Hsueh, YH., Tsai, PH., Lin, KS. et al. Antimicrobial effects of zero-valent iron nanoparticles on gram-positive Bacillus strains and gram-negative Escherichia coli strains. J Nanobiotechnol 15, 77 (2017). https://doi.org/10.1186/s12951-017-0314-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12951-017-0314-1