Abstract
Background
This study aimed to investigate a causal relationship between IBD and multiple kidney diseases using two-sample Mendelian randomization (MR) analyses.
Methods
We selected a group of single nucleotide polymorphisms (SNPs) specific to IBD as instrumental variables from a published genome-wide association study (GWAS) with 86,640 individuals of European ancestry. Summary statistics for multiple kidney diseases were obtained from the publicly available GWAS. Genetic data from one GWAS involving 210 extensive T-cell traits was used to estimate the mediating effect on specific kidney disease. Inverse-variance weighted method were used to evaluate the MR estimates for primary analysis.
Results
Genetic predisposition to IBD was associated with higher risk of IgA nephropathy (IgAN) (OR, 1.78; 95% CI, 1.45–2.19), but not membranous nephropathy, diabetic nephropathy, glomerulonephritis, nephrotic syndrome, chronic kidney disease, and urolithiasis. CD4 expression on CD4 + T cell had a significant genetic association with the risk of IgAN (OR, 2.72; 95% CI, 1.10–6.72). Additionally, consistent results were also observed when IBD was subclassified as ulcerative colitis (OR, 1.38; 95% CI, 1.10–1.71) and Crohn’s disease (OR, 1.37; 95% CI, 1.12–1.68). MR-PRESSO and the MR-Egger intercept did not identify pleiotropic SNPs.
Conclusions
This study provides genetic evidence supporting a positive casual association between IBD, including its subclassification as ulcerative colitis and Crohn’s disease, and the risk of IgAN. However, no casual association was found between IBD and other types of kidney diseases. Further exploration of IBD interventions as potential preventive measures for IgAN is warranted.
Similar content being viewed by others
Introduction
The inflammatory bowel diseases (IBD), including Crohn’s disease and ulcerative colitis, are also immune-mediated chronic or flares of inflammatory activity of the gastrointestinal tract [1]. Kidney-related extra-intestinal manifestations are common in IBD, occurring in 4 to 23% of IBD patients [2]. A cohort study involving renal biopsies of IBD patients with renal diseases showed that out of 896 IBD patients, 218 patients (24.3%) exhibited renal involvement, among which 25.7% were diagnosed with amyloidosis, 16.1% with immunoglobulin A nephropathy (IgAN), and 14.7% with crescentic glomerulonephritis [3]. Furthermore, a recent large-scale retrospective cohort study utilizing the United Kingdom electronic records database, identified 910 cases of chronic kidney disease (CKD) among 17,807 patients in the IBD cohort during follow-up, and found that IBD was associated with increased risk of CKD [4]. However, whether kidney diseases present as potential complication remains uncertain. Patients with IBD are susceptible to drug-related nephrotoxicity due to the nephrotoxic nature of certain medications used in their treatment, such as nonsteroidal anti-inflammatory drugs, methotrexate, and 5-aminosalicylic acid, and others [5, 6]. Distinguishing renal damage caused by nephrotoxic drugs or that resulting from the pathogenesis of IBD is challenging. Moreover, previous studies evaluating the association between IBD and kidney diseases had yielded conflicting results, likely due to the potential influence of residual confounding and reverse causation of observational studies [4, 7, 8]. Therefore, it’s essential to investigate whether there is a causal relationship between IBD and risk of renal diseases.
Mendelian randomization (MR) is an analytical approach used to infer causal associations between a modifiable exposure or risk factor and a clinically relevant outcome by integrating summary statistics from genome-wide association study (GWAS) [9, 10]. Based on the Mendel’s second law, genetic variants are randomly allocated in the population during gamete formation. The instrumental variables involved in the exposure will affect the outcomes proportionally if the exposure is causal. MR approach enables us to minimize the impact of confounding factors (e.g., environment) and reverse causation, serving as a mimic of the randomization in randomized controlled trial. This study conducted a two-sample MR analysis to examine the causal association between genetically predicted IBD and the risk of multiple kidney diseases. We aimed to evaluate which type of kidney disease primarily reflect this causal relationship.
Methods
Data sources
We used summary data on GWAS obtained from the International Inflammatory Bowel Disease Genetics Consortium (IIBDGC) [11]. It was a multi-ethnic large-scale genome-wide or Immunochip genotype data, in which we only included the extended cohort of 86,640 European individuals to identify new IBD risk loci and compared the genetic architecture of IBD susceptibility [11]. Beyond that, we also investigated the same consortium of Crohn’s disease and ulcerative colitis as subtypes of IBD with 20,550 and 17,647 cases vs. 41,642 and 47,179 controls, respectively. Regarding the multiple kidney diseases, including IgAN, membranous nephropathy, glomerulonephritis, diabetic nephropathy, nephrotic syndrome, chronic kidney disease, and kidney/ ureter/ bladder stone, the summary statistic were obtained from publicly available summary-level data of previously published GWAS and restricted to European descent [12,13,14]. We also retrieved the summary data from MR Base database from Medical Research Council Integrative Epidemiology Unit (MRC-IEU) and these datasets have already undergone the recommended quality control processes as previously described [16]. The extensive T-cell traits were derived from the SardiNIA project composed of GWAS data, which included 210 subtypes in the T-cell panel and cell marker expression levels on different T cells [15] The summary statistics could be archived in the IEU GWAS datasets (https://gwas.mrcieu.ac.uk). Table 1 shows the characteristics of all GWAS data for the exposure and outcome.
Assumptions for instrumental variable selection
The genetic instrumental variable selection used in MR analysis was performed based on following three fundamental assumptions [17, 18]. Firstly, selected instrumental variables should be powerfully associated with exposure. The F statistic was used to evaluate the strength of the relationship between instrumental variables and exposure [16]. The formula of F statistic is expressed as F = R2 (n-k-1)/[k× (1-R2)]. R2 refers to the cumulative explained variance of selected single-nucleotide polymorphisms (SNPs) on IBD and is defined as R2 = 2 × effect allele frequency (EAF) × (1-EAF) × b2. EAF is the effect allele frequency and b is the estimated genetic effect; n is the sample size; and k represent the number of selected instrumental variables. If the resulting F value > 10, it indicates a strong correlation between instrumental variables and exposure, thus minimizing bias caused by weak instrumental variables. Secondly, instrumental variables should be independent of confounders that influence both the exposure and outcome. Third, selected instrumental variables should affect outcomes only through exposure, and not via alternative pathways. This implies the absence of a horizontal pleiotropy effect between instrumental variables and outcome. MR-Egger regression was performed to identify potential horizontal pleiotropy pathways [17].
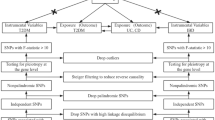
In the present study, SNPs significantly associated with IBD were selected as genetic instrumental variable, which met the F-statistic > 10. To avoid the potential bias caused by strong linkage disequilibrium (LD) among the selected instrumental variables, we performed LD clumping process using an r2 cutoff of 0.001 and a clumping distance = 10,000 kb. We systematically gathered information on effect allele (EA), EA frequency, effect sizes (β), S. E. and P-value for further analysis. To ensure data consistency, we harmonized the respective exposure and outcome datasets using effect allele frequencies. Additionally, we removing palindromic SNPs with minor allele frequency (MAF) > 0.01. Figure 1 displays a flowchart illustrating the whole procedure.
Statistical analysis
We conducted a two-sample MR in accordance with the Strengthening the Reporting of Observational Studies in Epidemiology-Mendelian Randomization (STROBE-MR) guidelines [19]. The causal effects of IBD on kidney diseases were estimated using the inverse-variance weighted (IVW) method for primary analysis. Moreover, to assess the robustness of IVW results, we compared them with other MR methods including MR-Egger, weighted median, simple mode, and weighted mode estimation. A consistent direction of estimates across all the MR methods increased confidence of causal relationship. The IVW method is essentially a meta-analysis method that requires all selected SNPs to be valid instrumental variables, with the intercept limited to zero. In the IVW method, a fixed-effect model was used when there was no heterogeneity (Cochrane’s Q P-value > 0.05), and a random-effect model was used otherwise. MR-Egger could be significantly influenced by outlying genetic variables, resulting in imprecise estimates, but it can still provide unbiased estimates even if there exist all invalid selected instrumental variables. The weighted median approach takes the median effect of all selected instrumental variables to provide accurate estimates with the prerequisite that at least half of SNPs are valid instrumental variables. The weighted mode approach remains still valid even if the other instrumental variables do not meet the requirements of MR method for causal inference. The MR-Egger regression was conducted to detect and adjusts for potential horizontal pleiotropic effects. Moreover, Cochran’s Q statistic was used to assess heterogeneity among the estimates from each SNP. The leave-one-out sensitivity analysis was performed to verify individual SNP that influenced the association disproportionately and to evaluate the stability of effect sizes. We also performed the Mendelian randomization pleiotropy residual sum and outlier (MR-PRESSO) test to estimate and correct outliers of instrumental variables with horizontal pleiotropic effects. Post-hoc power calculation for the main analyses was performed using mRnd website (http://cnsgenomics.com/shiny/mRnd/) [20]. Statistical analyses were conducted in R software (version 4.0.2; using the “TwoSampleMR”, “MendelianRandomization” and “MR-PRESSO” R package; R Foundation for Statistical Computing, Vienna, Austria). Two-tailed P < 0.05 was considered statistically significant.
Results
Genetic variants selection
The current study evaluated the causal effect of genetically predicted IBD on multiple kidney diseases. After removing SNPs with LD (r2 > 0.001 within 10,000 kb), palindromic SNPs (P > 5 × 108 and F < 10), and duplicated SNPs, we finally selected 67 to 129 instrumental SNPs for each trait. Among all selected SNPs, 67 independent and significant SNPs were associated with IgAN (cases = 977), 108 associated with membranous nephropathy (cases = 2,150), 129 associated with glomerulonephritis (cases = 4,613), 79 associated with nephrotic syndrome (cases = 480), 129 associated with diabetic nephropathy (cases = 3,283), 82 associated with CKD (cases = 12,385), and 120 associated with urolithiasis (cases = 3,625). Detailed information on the genetic variants, including SNPs, EA, EA frequency, effect sizes on IBD, ulcerative colitis, and Crohn’s disease and multiple kidney diseases is provided in Additional file 1: Table S1–S9.
Estimates of causal effect of IBD on kidney diseases
The results of IVW with a fixed-effect model estimates showed a positive correlation between the genetically predicted IBD and the risk of IgAN [odds ratio (OR) 1.78 (95% confidence interval (CI) 1.45–2.19), P < 0.001]. However, no significant genetic association was detected between IBD and risk of membranous nephropathy (OR, 0.93; 95%CI, 0.79–1.09), glomerulonephritis (OR, 1.01; 95%CI, 0.96–1.06), diabetic nephropathy (OR, 1.01; 95%CI, 1.00–1.02), nephrotic syndrome (OR, 1.04; 95%CI, 0.92–1.19), chronic kidney disease (OR, 1.02; 95%CI, 0.98–1.07), and urolithiasis (OR, 1.03; 95%CI, 0.98–1.09) (Fig. 2).
Estimates of causal effect of IBD and subclassification on IgAN
We further compared with other MR methods to evaluate the robustness of IVW results of causal effect of IBD and subclassification on IgAN. The MR estimates of weighted median [OR 1.82 (95% CI 1.33–2.49), P < 0.001] and weighted mode methods [OR 1.91 (95% CI 1.16–3.14), P = 0.01] also showed consistent results (Table 2; Fig. 3A and B). Post hoc power calculations indicated that the study had sufficient statistical powered (.100%) to detect an assuming of the real causal OR of 1.78 for the risk of IgAN with a total sample size of 5957 (19.6% ratio of IgAN case to control) and the significance level α of 0.05.
The genetic changes in the subclassification of IBD as ulcerative colitis, were also positively correlated with the risk of IgAN, which were showed in IVW based on a fixed-effect model [OR 1.38 (95% CI 1.10–1.71), P = 0.005; Additional file 1: Table S10, Additional file 1: Fig.S1A–S1B]. The results of IVW with a random-effect model showed that the genetic changes in Crohn’s disease were positively correlated with the risk of IgAN [OR 1.37 (95% CI 1.12–1.68), P = 0.002; Additional file 1: Table S11, Additional file 1: Fig.S2A–S2B]. However, no significant causal association between ulcerative colitis or Crohn’s disease and IBD were found in MR-Egger, weighted median and weighted mode methods (Additional file 1: Table S10–S11).
Estimates of causal association mediated by T cells traits and IgAN
The detailed information regarding the MR result, including EA, EA frequency, effect sizes on specifical T-cell traits and IgAN is shown in Additional file 1: Table S12. After harmonizing the respective T-cell phenotype exposure and outcome datasets, we identified 42 out of a total of 210 T-cell phenotypes had valid instrumental variables. Subsequently analyzing the 42 T-cell phenotypes in MR analysis, we only observed a significant genetic association between CD4 expression on CD4 + T cells and IgAN [OR 2.72 (95% CI 1.10–6.72), P = 0.029]. No statistically significant differences were observed for other T-cell traits (Additional file 1: Table S13).
Sensitivity analyses
Sensitivity analyses using the leave-one-out associations approach suggested that the causal association was not biased by potential driving SNP (Fig. 3C, Additional file 1: Fig.S1C, and Additional file 1: Fig. S2C). The horizontal pleiotropy between instrumental variables and outcome was assessed by MR-Egger regression, and the results indicated the predicted effect sizes of IBD, ulcerative colitis, and Crohn’s disease on IgAN were comparable and consistent in the direction [IBD: intercept = 0.0112 (SE = 0.031), P = 0.75, Fig. 3B], [ulcerative colitis: intercept = -0.007 (SE = 0.04), P = 0.87, Additional file 1: Fig.S1B], and [Crohn’s disease: intercept = -0.009 (SE = 0.038), P = 0.82, Additional file 1: Fig.S2B]. Besides, the MR-PRESSO method did not detect outlying SNPs causing horizontal pleiotropy [P-value for MR-PRESSO global test = 0.67 (IBD), 0.99 (ulcerative colitis), and 0.86 (Crohn’s disease)]. Furthermore, no heterogeneity of IBD and ulcerative colitis on IgAN was found in IVW and MR Egger method using the Cochran’s Q test [Cochran’s Q = 62.958, P = 0.53 (MR-Egger) and P = 0.56 (IVW), Fig. 3D] and [Cochran’s Q = 45.04, P = 0.10 (MR-Egger) and P = 0.17 (IVW), Additional file 1: Fig.S1D]. However, there was significant heterogeneity in Crohn’s disease [Cochran’s Q = 93.60, P = 0.01 (MR-Egger) and P = 0.02 (IVW), Additional file 1: Fig.S2D].
Discussion
The current study revealed a positive correlation between IBD and the risk of IgAN, and CD4 expression on CD4 + T cell could involve the causal effects. Additionally, the positive associations were also observed when subclassifying IBD into ulcerative colitis and Crohn’s disease. However, no statistical differences were found between IBD and risk of membranous nephropathy, glomerulonephritis, diabetic nephropathy, nephrotic syndrome, chronic kidney disease, and urolithiasis.
A observational study evaluated 83 kidney biopsy specimens from patients with IBD and found that IgAN was the most prevalent diagnosis (24%). This prevalence was significantly higher compared to all native non-IBD kidney biopsy specimens (24% versus 8%) [21]. Another retrospective analysis of renal biopsies done for IBD patients in Egypt revealed that out of 896 IBD patients, 218 (24.3%) developed renal complications, with IgAN being the second most common renal pathological diagnosis (16.1%) [3]. Additionally, a recent population-based cohort study including 3963 biopsy-verified IgAN patients and 19,978 matched controls, confirmed a correlation between IgAN and IBD, and further that a diagnosis of IBD among patients with IgAN were more likely than to progress into end-stage kidney disease than those without IBD [22]. To our knowledge, apart from the aforementioned cohort studies [3, 21, 22], the association between IgAN and IBD has mainly been described through single earlier case reports [23,24,25,26]. In present MR analysis, the use of SNPs as proxies of IBD allowed us to minimize the impact of confounding factors and inverse causality, which are common in observational study [27]. Our study provides genetic-level support for the positive association between IBD and the risk of IgAN, but not other types of kidney diseases. This finding aligns with the recent announcement by the Unite State Food and drug administration (FDA) and clinical trials that budesonide in oral capsules, designed to release active compounds upon reaching the distal ileum, is primarily intended for the treatment of adult primary IgA nephropathy, rather than other types of kidney diseases [28,29,30].The underlying mechanisms involved in deposition of IgA in glomerular of patients with inflammation of the intestinal mucosa remain to be unraveled. Recent advances have highlighted the important role of the gut-kidney axis in pathophysiologic connection between IBD and IgAN [31,32,33]. Firstly, it has been observed that most IBD patients experience hyperactivity of IgA1-secreting cells in the lesions of intestinal mucosa, which leads to an imbalance in the of immunoglobulin production [4, 34]. This imbalance could be due to the impair of mucosal integrity, which acts as a barrier to antigenic stimulation. Additionally, an increase in intestinal permeability and an inadequate response to the microbiota (rather than specific taxa), potentially driven by host cytokines like B-cell activating factor, such as B-cell activating factor [33]. Furthermore, recent GWAS have identified 9 genetic variants that control composition of gut microbiota may be associated with susceptibility to IgAN. This finding suggests that host genetics can influence gut microbiota in IgAN [35]. Interestingly, our results indicated a positive correlation between CD4 expression on CD4 + T cell and the risk of IgAN, among 210 extensive T-cell traits. Previous study have shown that serum levels of IgA strongly correlate with the percentage of CD4 + CD45RO + cells in peripheral blood in patients with IgAN, indicating that CD4 + T cells might also play a role in abnormal IgA1 glycosylation process [36,37,38]. Therefore, it might be worth exploring clinical trials with drugs that target the imbalance in the CD4 + T cell compartment.
Limitations
This study has several limitations. First, we cannot determine the extent of overlapping participants involved between data sources for SNPs-exposure and outcome consortia in two sample MR analyses. However, to minimize the potential deviation from participant overlap, we employed F statistic and instrumental variables filtering procedures. Second, all analyses were based from European ancestry individuals, and it is necessary to conduct further GWAS studies in other ethnicities to validate the results. Third, we investigated the casual effect of only one T-cell traits on the risk of IgAN, and it is possible that there were insufficient numbers of SNPs available to determine the causal effect. Hence, caution is needed when interpreting the results. Finally, we were unable to completely exclude the possibility of other direct causal pathway from the IBD-predisposing genetic variants to IgAN. To note, the subgroup analyses demonstrated a significant heterogeneity within the population with Crohn’s disease. This suggests the presence of potential variations in the validity of SNPs and false positive results, which can be attributed to the constraint imposed by the limited sample size. Nevertheless, both the MR-Egger intercept and MR-PRESSO analyses showed little heterogeneity among instrumental variables of IBD and minimal directional horizontal pleiotropy, suggesting a lower likelihood of bias.
Conclusions
The current MR study provides robust evidence supporting the causal association between genetic susceptibility to IBD and an increased risk of IgAN, as well as its specific subtypes including ulcerative colitis and Crohn’s disease. However, there is insufficient evidence to establish a causal link between IBD and other kidney diseases such as, membranous nephropathy, glomerulonephritis, diabetic nephropathy, nephrotic syndrome, chronic kidney disease, and urolithiasis. Further investigation using updated data from large-scale genetic studies is necessary to validate and reinforce the findings.
Data Availability
Only publicly available data were used in this study, and data sources and handling of these data were described in the Materials and Methods and supplementary files. The datasets analyzed for this study are available on the website (https://gwas.mrcieu.ac.uk). Further inquiries can be directed to the corresponding author Wei Chen.
Abbreviations
- IgAN:
-
Immunoglobulin A nephropathy
- ESKD:
-
End-stage kidney disease
- IBD:
-
Inflammatory bowel disease
- MR:
-
Mendelian randomization
- GWAS:
-
Genome-wide association study
- SNPs:
-
Single-nucleotide polymorphisms
- EAF:
-
Effect allele frequency
- MAF:
-
Minor allele frequency
- LD:
-
Minor allele frequency
- EA:
-
Effect allele
- IVW:
-
Inverse-variance weighted
- MR-PRESSO:
-
Mendelian randomization pleiotropy residual sum and outlier
- OR:
-
Odds ratio
- CI:
-
Confidence interval
References
Chang JT. Pathophysiology of Inflammatory Bowel Diseases. N Engl J Med. 2020;383(27):2652–64.
Pardi DS, Tremaine WJ, Sandborn WJ, McCarthy JT. Renal and urologic complications of inflammatory bowel disease. Am J Gastroenterol. 1998;93(4):504–14.
Elaziz MMA, Fayed A. Patterns of renal involvement in a cohort of patients with inflammatory bowel disease in Egypt. Acta Gastroenterol Belg. 2018;81(3):381–5.
Vajravelu RK, Copelovitch L, Osterman MT, Scott FI, Mamtani R, Lewis JD, et al. Inflammatory Bowel Diseases are Associated with an increased risk for chronic kidney Disease, which decreases with age. Clin Gastroenterol Hepatol. 2020;18(10):2262–8.
Achit H, Peyrin-Biroulet L, Ayav C, Guillemin F, Frimat L. Lack of monitoring is Associated with risk of Acute kidney events among patients with inflammatory bowel disease. J Clin Med. 2022;11(11).
Guillo L, Delanaye P, Flamant M, Figueres L, Karam S, Lemoine S, et al. Kidney function monitoring in inflammatory bowel disease: the MONITORED consensus. Dig Liver Dis. 2022;54(3):309–15.
Primas C, Novacek G, Schweiger K, Mayer A, Eser A, Papay P, et al. Renal insufficiency in IBD–prevalence and possible pathogenetic aspects. J Crohns Colitis. 2013;7(12):e630–4.
Lewis B, Mukewar S, Lopez R, Brzezinski A, Hall P, Shen B. Frequency and risk factors of renal insufficiency in inflammatory bowel disease inpatients. Inflamm Bowel Dis. 2013;19(9):1846–51.
Emdin CA, Khera AV, Kathiresan S. Mendelian Randomization JAMA. 2017;318(19):1925–6.
Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian randomization as an Approach to assess causality using Observational Data. J Am Soc Nephrol. 2016;27(11):3253–65.
Liu JZ, van Sommeren S, Huang H, Ng SC, Alberts R, Takahashi A, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47(9):979–86.
Xie J, Liu L, Mladkova N, Li Y, Ren H, Wang W, et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat Commun. 2020;11(1):1600.
Feehally J, Farrall M, Boland A, Gale DP, Gut I, Heath S, et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21(10):1791–7.
Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, et al. Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun. 2016;7:10023.
Orrù V, Steri M, Sidore C, Marongiu M, Serra V, Olla S, et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat Genet. 2020;52(10):1036–45.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some Invalid Instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304–14.
Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, et al. Strengthening the reporting of Observational Studies in Epidemiology using mendelian randomization: the STROBE-MR Statement. JAMA. 2021;326(16):1614–21.
Brion M-JA, Shakhbazov K, Visscher PM. Calculating statistical power in mendelian randomization studies. Int J Epidemiol. 2013;42(5):1497–501.
Ambruzs JM, Walker PD, Larsen CP. The histopathologic spectrum of kidney biopsies in patients with inflammatory bowel disease. Clin J Am Soc Nephrol. 2014;9(2):265–70.
Rehnberg J, Symreng A, Ludvigsson JF, Emilsson L. Inflammatory bowel disease is more common in patients with IgA nephropathy and predicts progression of ESKD: A Swedish Population-Based Cohort Study. J Am Soc Nephrol. 2021;32(2):411–23.
Terasaka T, Uchida HA, Umebayashi R, Tsukamoto K, Tanaka K, Kitagawa M, et al. The possible involvement of intestine-derived IgA1: a case of IgA nephropathy associated with Crohn’s disease. BMC Nephrol. 2016;17(1):122.
Choi J-Y, Yu CH, Jung H-Y, Jung MK, Kim Y-J, Cho J-H, et al. A case of rapidly progressive IgA nephropathy in a patient with exacerbation of Crohn’s disease. BMC Nephrol. 2012;13:84.
Trimarchi HM, Iotti A, Iotti R, Freixas EA, Peters R. Immunoglobulin a nephropathy and ulcerative colitis. A focus on their pathogenesis. Am J Nephrol. 2001;21(5):400–5.
de Moura CG, de Moura TGG, de Souza SP, Testagrossa L. Inflammatory bowel disease, ankylosing spondylitis, and IgA nephropathy. J Clin Rheumatol. 2006;12(2):106–7.
Davey Smith G, Hemani G. Mendelian randomization: genetic anchors for causal inference in epidemiological studies. Hum Mol Genet. 2014;23(R1):R89–98.
Fellström BC, Barratt J, Cook H, Coppo R, Feehally J, de Fijter JW, et al. Targeted-release budesonide versus placebo in patients with IgA nephropathy (NEFIGAN): a double-blind, randomised, placebo-controlled phase 2b trial. Lancet. 2017;389(10084):2117–27.
Barratt J, Rovin BH, Cattran D, Floege J, Lafayette R, Tesar V, et al. Why target the gut to treat IgA nephropathy? Kidney Int Rep. 2020;5(10):1620–4.
Hastings MC, Bursac Z, Julian BA, Villa Baca E, Featherston J, Woodford SY, et al. Life expectancy for patients from the Southeastern United States with IgA Nephropathy. Kidney Int Rep. 2018;3(1):99–104.
Perše M, Večerić-Haler Ž. The role of IgA in the Pathogenesis of IgA Nephropathy. Int J Mol Sci. 2019;20(24):6199.
Monteiro RC, Berthelot L. Role of gut-kidney axis in renal diseases and IgA nephropathy. Curr Opin Gastroenterol. 2021;37(6):565–71.
Haniuda K, Gommerman JL, Reich HN. The microbiome and IgA nephropathy. Semin Immunopathol. 2021;43(5):649–56.
Kett K, Brandtzaeg P. Local IgA subclass alterations in ulcerative colitis and Crohn’s disease of the colon. Gut. 1987;28(8):1013–21.
He J-W, Zhou X-J, Li Y-F, Wang Y-N, Liu L-J, Shi S-F, et al. Associations of genetic variants contributing to Gut Microbiota Composition in Immunoglobin A Nephropathy. mSystems. 2021;6(1):e00819–20.
Sato M, Haizuka H, Asakura H. Relationship between serum IgA levels and CD4 subsets in IgA nephropathy. Nephron. 1994;68(1):20–4.
Ruszkowski J, Lisowska KA, Pindel M, Heleniak Z, Dębska-Ślizień A, Witkowski JM. T cells in IgA nephropathy: role in pathogenesis, clinical significance and potential therapeutic target. Clin Exp Nephrol. 2019;23(3):291–303.
Honkanen T, Mustonen J, Kainulainen H, Myllymäki J, Collin P, Hurme M, et al. Small bowel cyclooxygenase 2 (COX-2) expression in patients with IgA nephropathy. Kidney Int. 2005;67(6):2187–95.
Acknowledgements
We want to acknowledge the participants and investigators of the FinnGen study, UK.
Biobank, and other GWAS for providing the publicly available summary data used in this study.
Funding
This work was supported by grants from the National Natural Science Foundation of China (No. 81970599, 82170737); Key Laboratory of National Health Commission, and Key Laboratory of Nephrology, Guangdong Province, Guangzhou, China (Nos. 2002B60118 and 2020B1212060028); Guangzhou Key Discipline of Medicine (Geriatric Medicine, ZDXK202103).
Author information
Authors and Affiliations
Contributions
XJL, YQW and SYW designed the study and conducted the MR analysis. XHP and YHW did the literature research and contributed to the data collection. XJL, YQW and SYW drafted the manuscript. YYH and WC supervised the method and visualized the results. All authors contributed to the manuscript revision and read and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Ethical review and approval were waived for this study by Ethics Committee of Guangzhou First People’s Hospital because this study was conducted on summary-level genetic data from publicly available databases of large-scale genome-wide association studies.
Consent for publication
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Lian, X., Wang, Y., Wang, S. et al. Does inflammatory bowel disease promote kidney diseases: a mendelian randomization study with populations of European ancestry. BMC Med Genomics 16, 225 (2023). https://doi.org/10.1186/s12920-023-01644-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-023-01644-2