Abstract
Background
Elevated blood pressure is an important risk factor for cardiovascular disease and is also an important factor in global mortality. Military pilots are at high risk of cardiovascular disease because they undergo persistent noise, high mental tension, high altitude hypoxia, high acceleration and high calorie diet. Hypertension is the leading cause of cardiovascular disease in military pilots. In this study, we want to identify key genes from peripheral blood cells of military pilots with hypertension. Identification of these genes may help diagnose and control hypertension and extend flight career for military pilots.
Methods
We use RNA sequencing technology, bioinformatics analysis and Western blotting to identify key genes from peripheral blood cells of military pilots with hypertension.
Results
Our study detected 121 up-regulated genes and 623 down-regulated genes in the peripheral blood mononuclear cells (PBMCs) from hypertensive military pilots. We have also identified 8 important genes (NME4, PNPLA7, GGT5, PTGS2, IGF1R, NT5C2, ENTPD1 and PTEN), a number of gene ontology categories and biological pathways that may be associated with military pilot hypertension.
Conclusions
Our study may provide effective means for the prevention, diagnosis and treatment of hypertension for military pilot and extend their flight career.
Similar content being viewed by others
Background
Elevated blood pressure (BP) is an important risk factor of cardiovascular disease (CVD) and is also an important factor in global mortality, leading to about 9.4 million deaths per year [1]. The morbidity and mortality of CVD are associated with degrees of increased BP. For every increase of 20 mmHg in the systolic BP above 115 mmHg, the incidence of CVD risk will double [2]. In general, the threshold of hypertension is set to systolic pressure of 140 mmHg (150 mmHg for older adults) or diastolic pressure of 90 mmHg based on BP measurement in quiescent condition [3]. One-third American adults over 18 years old of age have hypertension, and 54% of old people (55- to 64-year-olds) have high BP. Among those over 75 years old of age in the United States, nearly 80% have hypertension [4].
Military pilots are at high risk of CVD because they are undergo persistent noise, high mental tension, high altitude hypoxia, high acceleration and high calorie diet [5]. Hypertension is the leading cause of CVD in military pilots. Although hypertension itself will not cause sudden disability in flight, but it is a main risk factor for disability in flight career and it is also one of the major reasons to cause the pilot grounded [6]. Wenzel et al. reported that the incidence of hypertension in Brazilian Air Force is about 22% [7]. Grossman et al. found 2.4% of the pilots had moderate or higher blood pressure in a 7.5-year follow-up study of Israeli Air Force pilots [8]. The hypertension prevalence rate was 9.7% in Chinese Air Force pilots, and the grounded rate of pilots was 21.7% among students in the flight academy because of hypertension or increased blood pressure [9]. Essential hypertension is a disease caused by complex, multifactorial and multigenic changes, and it is the result of both gene regulation and environmental impact [10]. In this study, we use RNA sequencing technology, bioinformatics analysis and Western blotting to identify key genes from peripheral blood cells of military pilots with hypertension. Identification of these genes may help diagnose and control hypertension and extend flight career for military pilots.
Methods
Study subject
For RNA-Seq, six samples of peripheral blood cell from military pilot (3 hypertensives and 3 normotensives) were collected. For quantitative RT-PCR and Western blotting analysis, another 8 samples of peripheral blood cell from fighter pilot (4 hypertensives and 4 normotensives) were collected. All samples collected with the help of doctors from Lintong Aviation Medical Evaluating and Training Center of Air Force, Xi’an, China. The average systolic blood pressure (SBP) of these hypertension pilots was above 160 mmHg and the average diastolic blood pressure (DBP) was above 100 mmHg. The average SBP of these normotensive pilots was below 135 mmHg and the average DBP was below 85 mmHg.
Peripheral blood mononuclear cells (PBMCs) isolation
Five milliliter whole blood collected was transferred to a 15 ml sterile centrifuge tube, 5 ml phosphate buffer saline (PBS) was added to dilute the whole blood. Five milliliter lymphocyte separation medium was added to another 15 ml sterile centrifuge tube. The diluted whole blood was transferred to the lymphocyte separation medium gently to avoid mixing. Then the blood was centrifuged at 2000 rpm for 20 min, room temperature. The white membrane cells of PBMSs were sucked into another sterile 15 ml centrifuge tube. Add 5 ml PBS and centrifuge at 1000 rpm for 10 min. Aspirate supernatant and add 2 ml ACK buffer to the tube, suspend the cells and keep standing for 5 min. Add 5 ml PBS and centrifuge at 1000 rpm for 10 min. Aspirate the supernatant, and the cells were quickly frozen in liquid nitrogen and used for further analysis.
RNA sequencing
RNA sequencing was entrusted to Novel Bioinformatics Co., Ltd., Shanghai, China. Using high-throughput Life technologies Ion Proton Sequencer, the transcript with poly(A)-containing RNA of Human were analyzed.
Quality control
Fast - QC software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was employed to evaluate the overall quality of sequencing data.
Gene expression calculation
The expression of genes is mainly calculated by RPKM (Reads Per Kb per Million reads) method. The formula is: \( RPKM=\frac{10^6C}{NL/{10}^3} \) RPKM is gene expression, C is the unique number of reads on the gene, and N is the unique number of total reads in the reference gene, and L is the base number of the gene coding region. RPKM method can eliminate the effects of gene length and sequencing on the expression of genes, which can be directly used to compare the gene expression differences between different samples.
Principal component analysis
We applied the PCA analysis based the whole gene expression table and the R script utilizing following package: MASS, evd, rgl and pvclust. Command we used was described as followings:
R script:
dat < − read.table(“all.rpkm.exp.txt”,sep = “\t”, header = T).
colnameall = colnames(dat).
colname = colnames(dat[,2:length(colnameall)]).
dat.pca < −princomp(dat[2:length(colnameall)]).
summary(dat.pca).
plot<−plot3d(dat.pca$loadings[,1],dat.pca$loadings[,2],dat.pca$loadings[,3],type = “s”,col. = col.,size = 0.8,xlab = “PC1”,ylab = “PC2”,zlab = “PC3”).
texts3d(dat.pca$loadings[,1],dat.pca$loadings[,2]-0.02,dat.pca$loadings[,3],texts = colname,font = 5).
GO analysis
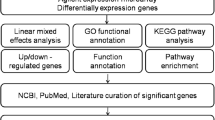
The difference of gene analysis based on the database from BP, MF, CC GO annotation in the three dimensions and all GOs were obtained. Each GO significance level was obtained by using the Fisher test (P Value) and so gene enrichment significant difference GOs were screened out [11, 12].
Pathway analysis
The differential expression genes filter out were annotated in KEGG database (http://www.genome.jp/kegg/) for Pathway annotations, and all the Pathway Terms of different genes involved were got. The Pathway of significance level is obtained by using the Fisher test (P Value), and the significant Pathway Term of differential expression gene enrichment was screened out.
Gene-act-network
The construction of gene interaction is to sort out the regulation of all the genes, and through the construction of signal transduction network, we could easily find the vein of gene signal transduction. Based on KEGG database, we could get gene interactions. So, we constructed gene and adjacency matrix using gene interactions [13,14,15,16,17].
Western blotting
PBMCs were isolated as before. Total protein was extracted by using the RIPA reagent (Beyotime, Shanghai, China). The sample protein concentration was tested by measured by BCA Protein Assay reagents (Thermo Scientific, Rockford, IL). After the protein electrophoresis, the samples were transferred to the PVDF membrane, which was then incubated for primary antibodies overnight at 4 °C, and then the membrane was incubated for horseradish peroxidase (HRP)-conjugated secondary antibodies for 2 h at room temperature. The primary antibodies used were as follows: NT5C2 rabbit polyclonal antibody (Proteintech, Wuhan, China), ENTPD1 rabbit polyclonal antibody (Proteintech, Wuhan, China), GGT5 rabbit polyclonal antibody (Proteintech, Wuhan, China), COX2 (PTGS2) rabbit polyclonal antibody (Proteintech, Wuhan, China), PTEN rabbit polyclonal antibody (Proteintech, Wuhan, China), IGF1R rabbit polyclonal antibody (Proteintech, Wuhan, China), rabbit anti-NME4 (Bioss Antibodies, Beijing, China), rabbit anti-PLCG2 (Bioss Antibodies, Beijing, China), rabbit anti-PI3 Kinase p110 delta (PIK3D) (Bioss Antibodies, Beijing, China), anti-PNPLA7 (Santa Cruz Biotechnology, Dallas, TX), anti-β-actin (Santa Cruz Biotechnology, Dallas, TX). The membrane was chemiluminescence using the chemiluminescent reagents (Millipore Corporation, Billerica, MA) and image-forming system Tanon 4200 (Tanon Science & Technology Co., Ltd., Shanghai, China).
Results
Overview of RNA sequencing data
We used Fast-QC online software (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to assess the quality of sequencing results (Results were not shown). The results showed that the sequencing data of all samples were qualified. Total raw reads of these six samples were about 12–15 million. GC content of these six samples was about 53%. The average reads mapped to human genome sequence were about 1.28 ± 0.78 × 107reads (96% of the total reads) in the six samples (Additional file 1: Table S1) and about 92% of the total reads (1.24 ± 0.74 × 107 reads) were mapped to human genome sequence uniquely. MapSplice was employed to map the reads. According to the mapping results, about 1 × 107 reads were mapped to the transcript exon, 7 × 106 reads were mapped to CDS, 2 × 106 reads were mapped to intron, 2 × 106 reads were mapped to the UTR regions and the rest reads (about 5% or less) were mapped to intergenic regions, TSS (transcription start site) and TES (transcription end site) (Fig. 1a). We also detected the distribution on chromosomes of these sample mapped reads (Fig. 1b). The results showed that the most reads were aligned to chromosome 1 (about 10% or more) and the least reads were aligned to chromosome Y (less than 0.2%).
Differential gene expression profiles between PBMCs from hypertensives and normotensives military pilots and GO analysis
The six samples were screened for difference gene expression with 3:3. The total number was 26, lower than the minimum standard of data analysis. Two discrete samples (Con4 and Hyp3) were deleted according to results of principal component analysis, and 744 differential genes were acquired. This number was enough for further analysis, so we adopted difference gene expression screened with 2:2.
To characterize the gene expression changes between hypertensives and normotensives military pilots’ PBMCs, differentially expressed genes were screened by the following standard: Log2FC > 0.585 or Log2FC < − 0.585, FDR < 0.05 and P value < 0.05. We found 744 differentially expressed genes between hypertensives and normotensives military pilots’ PBMCs. Of these genes, 121 genes were up-regulated in hypertensives military pilots’ PBMCs and 623 genes were down-regulated. PCA (principal components analysis) cluster analysis was used to compare differential expression of these two groups. The differential gene expression patterns of these samples were showed by gene thermal map (Fig. 2). Gene ontology analysis was used to seek the functions of these differentially expressed genes. The results showed that there were 337 genes belonged to protein binding and 64 genes belonged to ATP binding for molecular function (MF). There were 91 genes belonged to signal transduction and 70 genes belonged to small molecule metabolic process for biological process (BP). As for cellular component (CC), there were 311 genes belonged to membrane and 248 genes belonged to cytoplasm. Results from GO term analyzing showed that inflammatory response, protein binding and phagolysosome were the most significant for BP, MF and CC respectively (Fig. 3).
Gene Ontology (GO) Analysis of differentially expressed genes. GO analysis was annotated from three levels: BP, MF and CC. -Log10(P-value) was showed at abscissa axis and GO terms was showed at longitudinal axis. P value < 0.05 for all significant GO terms. BP: Biological Process; CC: Cellular Component; MF: Molecular Function
Pathways analysis of differential expression genes
The differential expression genes were involved in multiple GO, so we constructed functional relation network with significant GO-Term (p-value< 0.05) to reveal relationship between genes clearly based on hierarchical structure of GO (Additional file 2: Figure S1). Of these pathways, protein phosphorylation, toll-like receptor signaling pathway and cell surface receptor signaling pathway were in the core position (Additional file 2: Figure S1). Then, pathway-analysis was carried out to detect significant and important pathways of these differential expression genes. Influenza A and osteoclast differentiation were the most significant (Fig. 4a). Also, the top 20 of pathway enrichment was displayed in Fig. 4b. P-Value and gene number was indicated as circle size and color. Next, the pathways interaction network was built to analysis deeply. The analysis results showed that the most important pathways were apoptosis, Jak-STAT signaling pathway, toll-like receptor signaling and cytokine-cytokine receptor interaction (Additional file 3: Figure S2). Because these four pathways are located at the center of the all significant pathways and have the most arrowheads around, these four pathways are likely to be most important in the elevated blood pressure of military pilots. This result suggested that differential expression genes related to apoptosis, Jak-STAT signaling pathway, toll-like receptor signaling and cytokine-cytokine receptor interaction may have important role in the occurrence and development of elevated blood pressure of military pilots.
Pathway enrichment analysis of differentially expressed genes based on KEGG. a Histogram of pathway enrichment. -Log10(P-value) was showed at abscissa axis and pathway terms of KEGG was showed at longitudinal axis. b Top 20 of pathway enrichment was displayed by different colors and different sizes of the circle
Gene act network of differentially expressed genes
Although we got four important pathways related to elevated blood pressure of military pilots, we know that one gene may be involved in multiple signal transduction pathways at the same time. So next, we built gene act network based on the relationships between the differentially expressed genes including expression, binding, inhibition, activation and compound. This analysis method can form the corresponding regulation relationship between gene and gene and is easier to find important related genes under the intervention measures. By analysis the gene act network, we found that NME4, PNPLA7, GGT5, PTGS2, IGF1R, PLCG2, NT5C2, ENTPD1, PIK3CD and PTEN these ten genes were located at the center of the all significant genes and have the most arrowheads around (Fig. 5). And also, these ten genes were involved in apoptosis, Jak-STAT signaling pathway, toll-like receptor signaling and cytokine-cytokine receptor interaction signaling pathway previously mentioned. So Next, we will confirm changes of these genes in military pilots’ PBMCs of hypertensives and normotensives with Western blotting.
Validation of representative differentially expressed genes by western blotting
The expression of NME4, PNPLA7, GGT5, PTGS2, IGF1R, PLCG2, NT5C2, ENTPD1, PIK3CD and PTEN from PBMCs of hypertensive and normotensive military pilots were detected by Western blotting. The results from Western blotting showed that NME4 and PNPLA7 these two genes were up-regulated significantly and GGT5, PTGS2, IGF1R, NT5C2, ENTPD1 and PTEN these six genes were down-regulated significantly in PBMCs of military pilots with hypertension (Fig. 6). Although the expression change of PLCG2 and PIK3CD was not significant between hypertensives and normotensives, there was downward trend in PBMCs of hypertensive military pilots (Fig. 6). The results indicated that the expression change of NME4, PNPLA7, GGT5, PTGS2, IGF1R, NT5C2, ENTPD1 and PTEN could be as sign of elevation of blood pressure for military pilots.
Western blotting validation of relative expression levels of representative differentially expressed genes. a Western blotting of 10 representative differentially expressed genes from 4 hypertensive (H1, H2 H3 and H4) and 4 normotensive (N1, N2, N3, N4) military pilots. b Statistics of relative expression of Western blotting. Bars = means ± SD. *P < 0.05, **P < 0.01, NS, not significant, n = 4
Discussion
Military pilots are in a state of persistent noise, high mental tension, high altitude hypoxia, high acceleration and their high calorie diet, so hypertension is a very common disease in this group. To identify the key genes that related to hypertension of military pilots is very necessary for prevention, diagnosis and treatment of this disorder. In this study, we used RNA sequencing of PBMCs to identify differential gene expression profile between the hypertensive and normotensive military pilots.
Six samples were sequenced and the difference gene expression was compared with 3:3. Because two samples (Con4 and Hyp3) were discrete, so we took them out and adopted different gene expression screened with 2:2. 121 up-regulated genes and 623 down-regulated genes were identified as different expressed genes between hypertensive and normotensive military pilots. We selected 10 important and significant genes according to results of gene act network analysis. Western blotting was employed to validate the expression change of the 10 genes above. The results showed that expression change of NME4, PNPLA7, GGT5, PTGS2, IGF1R, NT5C2, ENTPD1 and PTEN were consistent with the results of RNA sequencing.
NME4 (also known as NDPK-D, Nm23-H4) belongs to the Nm23 family, which includes 10 isoforms (NME1 to NME10) [18]. The classical function of NME4 as a group I isoform is its NDP kinase activity [19]. PNPLA7 (also known as NTE-R1 or NRE) is a member of the PNPLAs family, and its encoded protein is very conservative in mice, rats and human. PNPLA7 plays an important role in the hydrolysis of triglycerides, energy metabolism, lipid formation and adipocyte differentiation [20]. Up-regulation of PNPLA7 indicates elevated blood lipids, which has some correlation with hypertension.
The main expression of Gamma-glutamyl transferase 5 (GGT5) is on the surface of cell membrane, and the role of GGT5 is to hydrolyze the gamma-glutamyl bond glutathione [21]. There is no phenotypic abnormality in GGT5 knockout mice under normal conditions. But GGT5 gene knockout mice were unable to metabolize LTC4, resulting in diminished potential of neutrophils infiltrating into the peritoneum. The expression and function of GGT5 in human are rarely reported [22]. PTGS2(also known as COX-2) could promote carcinogenesis and metastasis of multiple types of tumors. Expression of PTGS2 could be dramatically up-regulated by high levels of noise exposure and high altitude hypoxia [23, 24]. Park et al. reported that high fat diet could reduce PTGS2 expression [25], indicating that expression change of PTGS2 may be caused by noise exposure, high altitude hypoxia and high fat diet. IGFR is a member of the receptor tyrosine family and can form homodimers with insulin receptor (InsR) to identify and bind to the ligand of insulin receptor IGF1 and IGF2 [26]. Heterozygous deficiency of Igf1r reduced postnatal growth and develop age-dependent insulin resistance. Old-aged Igf1r+/− mice had increased adiposity and exhibited increased adipogenesis [27], indicated that reduced expression of IGF1R may have a correlation with hypertension. NT5C2 plays an important role in purine metabolism. Some papers reported that there were some relationships between somatic mutations of NT5C2 and T-acute lymphoblastic leukemias (T-ALL) [28, 29]. ENTPD1 (also known as CD39) is a plasma membrane protein and its role is to hydrolyze extracellular ATP and ADP to AMP. Helenius et al. reported that the expression of ENTPD1 was significantly reduced in small arterial endothelial cells in patients with pulmonary arterial hypertension (PAH). The attenuation function of ENTPD1 is closely related to vascular dysfunction, suggested that ENTPD1 may be a novel target for PAH therapy [30]. Their results strongly suggested that down-regulation of ENTPD1 may be a sign of hypertension, consistent with our results. PTEN signaling pathway is one of the most important signaling pathways that regulate individual development, and participate in the process of cell proliferation, differentiation, aging and apoptosis. Schwerd et al. reported that mutation of PTEN led severe macrocephaly and mild intellectual disability in adolescent [31]. Skalska-Sadowska et al. reported that mutations of PTEN induced T-ALL [32]. These results suggested that abnormal expression of PTEN could lead nervous system abnormalities and hematologic disease, maybe have some relationship with hypertension.
Some of the above eight genes are associated with a high-fat diet, some are associated with noise and high-altitude hypoxia, some may be associated with high mental tension. We hypothesized that the expression change of these genes may affect the level of insulin-like growth factor and small vessel remodeling to induce hypertension under the long-term impact of flight environment. Therefore, we should pay attention to the expression changes of these eight genes in the prevention and control of military pilots’ hypertension. Early prevention and early treatment according to the expression change of these eight genes, may extend flight career of military pilots.
At last, we acknowledge that the lack of non-pilot controls is a limitation of the study. The eight differential genes we identified may not be pilots specific. Therefore, our results may lead to some errors in the prevention, diagnosis and treatment of pilot hypertension. But our study still has good guidance for the prevention, diagnosis, and treatment of hypertension in pilots, because the samples used in RNA sequencing and Western blotting experiments in this study are all from military pilots. We will employ non-pilot controls including hypertensives and normotensives to identify differential genes more accurate in the future.
Conclusions
In summary, this study detected gene expression difference in PBMCs between military pilot hypertension and normotensives. We have also identified 8 important genes (NME4, PNPLA7, GGT5, PTGS2, IGF1R, NT5C2, ENTPD1 and PTEN), a few GO categories and biological pathways that may be associated with the military pilot hypertension. Our study may provide effective means for the prevention, diagnosis and treatment hypertension of military pilot, extend their flight career.
Abbreviations
- BP:
-
Blood pressure
- CC:
-
Cellular component
- CDS:
-
Coding sequence
- CVD:
-
Cardiovascular disease
- DBP:
-
Diastolic blood pressure
- FDR:
-
False discovery rate
- GO:
-
Gene ontology
- HRP:
-
Horseradish peroxidase
- MF:
-
Molecular function
- PBMCs:
-
Peripheral Blood Mononuclear Cells
- PBS:
-
Phosphate buffer saline
- PVDF:
-
Polyvinylidene fluoride
- RPKM:
-
Reads per Kb per million reads
- SBP:
-
Systolic blood pressure
- T-ALL:
-
T-acute lymphoblastic leukemias
- TES:
-
Transcription end site
- TSS:
-
Transcription start site
References
Lim SS, Vos T, Flaxman AD, Danaei G, Shibuya K, Adair-Rohani H, Amann M, Anderson HR, Andrews KG, Aryee M, et al. A comparative risk assessment of burden of disease and injury attributable to 67 risk factors and risk factor clusters in 21 regions, 1990-2010: a systematic analysis for the global burden of disease study 2010. Lancet. 2012;380(9859):2224–60.
Lewington S, Clarke R, Qizilbash N, Peto R, Collins R. Age-specific relevance of usual blood pressure to vascular mortality: a meta-analysis of individual data for one million adults in 61 prospective studies. Lancet. 2002;360(9349):1903–13.
Weber MA, Schiffrin EL, White WB, Mann S, Lindholm LH, Kenerson JG, Flack JM, Carter BL, Materson BJ, Ram CV, et al. Clinical practice guidelines for the management of hypertension in the community a statement by the American Society of Hypertension and the International Society of Hypertension. J Hypertens. 2014;32(1):3–15.
Mozaffarian D, Benjamin EJ, Go AS, Arnett DK, Blaha MJ, Cushman M, Das SR, de Ferranti S, Despres JP, Fullerton HJ, et al. Executive summary: heart disease and stroke statistics--2016 update: a report from the American Heart Association. Circulation. 2016;133(4):447–54.
Shamiss A, Meisel S, Rosenthal T. Acute hypertensive response to +Gz acceleration in mildly hypertensive pilots. Aviat Space Environ Med. 1993;64(8):751–4.
McCrary BF, Van Syoc DL. Permanent flying disqualifications of USAF pilots and navigators (1995-1999). Aviat Space Environ Med. 2002;73(11):1117–21.
Wenzel D, Souza JM, Souza SB. Prevalence of arterial hypertension in young military personnel and associated factors. Rev Saude Publica. 2009;43(5):789–95.
Grossman C, Grossman A, Koren-Morag N, Azaria B, Goldstein L, Grossman E. Interventricular septum thickness predicts future systolic hypertension in young healthy pilots. Hypertens Res. 2008;31(1):15–20.
Quan SZ, Ma HY, Luo D, Dong L, Zhou JL, Zhu MC. Dependability research on hypertension and risk factors of cardiovascular disease in pilots. Chin J Clinicians (Electronic Version). 2012;6(4):68–71.
Korkor MT, Meng FB, Xing SY, Zhang MC, Guo JR, Zhu XX, Yang P. Microarray analysis of differential gene expression profile in peripheral blood cells of patients with human essential hypertension. Int J Med Sci. 2011;8(2):168–79.
The Gene Ontology (GO) project in. Nucleic. Acids Res. 2006;34(Database issue):D322–6.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9.
Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12(1):37–46.
Li C, Li H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics. 2008;24(9):1175–82.
Wei Z, Li H. A Markov random field model for network-based analysis of genomic data. Bioinformatics. 2007;23(12):1537–44.
Zhang JD, Wiemann S. KEGGgraph: a graph approach to KEGG PATHWAY in R and bioconductor. Bioinformatics. 2009;25(11):1470–1.
Spirin V, Mirny LA. Protein complexes and functional modules in molecular networks. Proc Natl Acad Sci U S A. 2003;100(21):12123–8.
Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D, Huang Z, Jiang J, Yanamala N, Seffouh A, et al. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. J Biol Chem. 2013;288(1):111–21.
Schlattner U, Tokarska-Schlattner M, Epand RM, Boissan M, Lacombe ML, Klein-Seetharaman J, Kagan VE. Mitochondrial NM23-H4/NDPK-D: a bifunctional nanoswitch for bioenergetics and lipid signaling. Naunyn Schmiedeberg’s Arch Pharmacol. 2015;388(2):271–8.
Zhang X, Zhang J, Wang R, Guo S, Zhang H, Ma Y, Liu Q, Chu H, Xu X, Zhang Y, et al. Hypermethylation reduces the expression of PNPLA7 in hepatocellular carcinoma. Oncol Lett. 2016;12(1):670–4.
Wickham S, West MB, Cook PF, Hanigan MH. Gamma-glutamyl compounds: substrate specificity of gamma-glutamyl transpeptidase enzymes. Anal Biochem. 2011;414(2):208–14.
Hanigan MH, Gillies EM, Wickham S, Wakeham N, Wirsig-Wiechmann CR. Immunolabeling of gamma-glutamyl transferase 5 in normal human tissues reveals that expression and localization differ from gamma-glutamyl transferase 1. Histochem Cell Biol. 2015;143(5):505–15.
Faoro V, Fink B, Taudorf S, Dehnert C, Berger MM, Swenson ER, Bailey DM, Bartsch P, Mairbaurl H. Acute in vitro hypoxia and high-altitude (4,559 m) exposure decreases leukocyte oxygen consumption. Am J Physiol Regul Integr Comp Physiol. 2011;300(1):R32–9.
Sun Y, Yu J, Lin X, Tang W. Inhibition of cyclooxygenase-2 by NS398 attenuates noise-induced hearing loss in mice. Sci Rep. 2016;6:22573.
Park S, Park NY, Valacchi G, Lim Y. Calorie restriction with a high-fat diet effectively attenuated inflammatory response and oxidative stress-related markers in obese tissues of the high diet fed rats. Mediat Inflamm. 2012;2012:984643.
Ii M, Li H, Adachi Y, Yamamoto H, Ohashi H, Taniguchi H, Arimura Y, Carbone DP, Imai K, Shinomura Y. The efficacy of IGF-I receptor monoclonal antibody against human gastrointestinal carcinomas is independent of k-ras mutation status. Clin Cancer Res. 2011;17(15):5048–59.
Thakur S, Garg N, Zhang N, Hussey SE, Musi N, Adamo ML. IGF-1 receptor haploinsufficiency leads to age-dependent development of metabolic syndrome. Biochem Biophys Res Commun. 2017;486(4):937–44.
Meyer JA, Wang J, Hogan LE, Yang JJ, Dandekar S, Patel JP, Tang Z, Zumbo P, Li S, Zavadil J, et al. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat Genet. 2013;45(3):290–4.
Tzoneva G, Perez-Garcia A, Carpenter Z, Khiabanian H, Tosello V, Allegretta M, Paietta E, Racevskis J, Rowe JM, Tallman MS, et al. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat Med. 2013;19(3):368–71.
Helenius MH, Vattulainen S, Orcholski M, Aho J, Komulainen A, Taimen P, Wang L, de Jesus Perez VA, Koskenvuo JW, Alastalo TP. Suppression of endothelial CD39/ENTPD1 is associated with pulmonary vascular remodeling in pulmonary arterial hypertension. Am J Physiol Lung Cell Mol Physiol. 2015;308(10):L1046–57.
Schwerd T, Khaled AV, Schurmann M, Chen H, Handel N, Reis A, Gillessen-Kaesbach G, Uhlig HH, Abou Jamra R. A recessive form of extreme macrocephaly and mild intellectual disability complements the spectrum of PTEN hamartoma tumour syndrome. Eur J Hum Genet. 2016;24(6):889–94.
Skalska-Sadowska J, Dawidowska M, Szarzynska-Zawadzka B, Jarmuz-Szymczak M, Czerwinska-Rybak J, Machowska L, Derwich K. Translocation t(8;14)(q24;q11) with concurrent PTEN alterations and deletions of STIL/TAL1 and CDKN2A/B in a pediatric case of acute T-lymphoblastic leukemia: a genetic profile associated with adverse prognosis. Pediatr Blood Cancer. 2017;64(4)
Acknowledgements
We are indebted to the members of our laboratories at the Department of Aerospace Physiology, Fourth Military Medical University.
Funding
This work was supported by the grant of Youth Training Project of PLA Medical Science and Technology, Funded by the General Logistics Department of PLA, China (Grant No:15QNP066). The cost of some reagents of this study was supported by Natural Science Foundation of Shaanxi, China (Grant No:2016JQ8002; 2012JQ4001) and Natural Science Foundation of China (Grant No:31300979). The publication cost of this article was sponsored by Youth Training Project of PLA Medical Science and Technology, China (Grant No:15QNP066).
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
XCZ and ZBY conceived and designed the study. XCZ, SHY, YQY, LZ, BJ and SJ performed the experiments. SHY and XZ collected the blood sample. YQY, LZ and SJ extracted and quantified protein sample. XCZ, SHY, XZ and BJ did Western blotting and data analysis. XCZ, YQY, LZ and SJ analyzed the sequencing data. XCZ interpreted the data and wrote the paper. ZBY revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Ethics approval of this study was obtained from committee of Xijing Hospital and all procedures performed involving human participants were in accordance with the ethical standards of this committee. Informed consent was obtained from all individual participants as verbal form included in the study. All individual participants agreed to choose verbal form instead of written form and this was approved by the committee of Xijing Hospital.
Consent for publication
Not applicable.
Competing interests
The authors declared that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
Table S1. Statistics of raw and mapped reads from RNA-seq analysis of PBMCs from hypertensives (Hyp) and normotensives (Con) military pilot. (PPTX 62 kb)
Additional file 2:
Figure S1. Relationship between genes based on hierarchical structure of GO. (PPTX 664 kb)
Additional file 3:
Figure S2. Pathways interaction network analysis. (PPTX 496 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Zhao, XC., Yang, SH., Yan, YQ. et al. Identification of differential gene expression profile from peripheral blood cells of military pilots with hypertension by RNA sequencing analysis. BMC Med Genomics 11, 59 (2018). https://doi.org/10.1186/s12920-018-0378-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12920-018-0378-2