Abstract
Background
TP53 is the most frequently mutated gene in human cancers. Previous studies reported that TP53 mutations correlated with poor prognoses in patients with head and neck squamous cell carcinoma (HNSCC). However, the relationship between TP53 mutations and hypopharyngeal squamous cell carcinoma (HPSCC) is not known. The current study aimed to evaluate TP53 mutation status as a predictive biomarker in patients with HPSCC.
Methods
We retrospectively reviewed the clinical charts of 57 HPSCC patients treated with initial surgery between 2008 and 2014. TP53 mutation status was determined by Sanger sequencing, and patients were classified into wild-type, missense mutation, and truncating mutation groups. Additionally, p53 expression was determined using immunohistochemistry in surgical specimens.
Results
TP53 mutations were identified in 39 (68%) patients. The 3-year disease-specific survival (DSS) rate of wild-type, missense mutation, and truncating mutation group were 94%, 61%, and 43%, respectively. The TP53 mutation group displayed significantly worse DSS and overall survival rates than the wild-type group (P = 0.01 and P = 0.007, respectively). Multivariate analyses revealed that the presence of TP53 mutations and ≥4 metastatic lymph nodes were independent adverse prognostic factors for HPSCC. p53 immunopositivity was detected in 22 patients, including 5 (28%) and 17 (71%) patients in the wild-type and missense mutation groups, whereas none of the patients with truncating mutation exhibited p53 immunopositivity (P = 0.0001).
Conclusion
The TP53 mutation status correlated with poor prognosis in surgically treated HPSCC patients. Specifically, truncating mutations which were not detected by p53 immunohistochemistry were predictive of worst survival.
Similar content being viewed by others
Background
Among squamous cell carcinomas (SCC) originating in the upper aerodigestive tract, the management of hypopharyngeal squamous cell carcinoma (HPSCC) remains to be one of the most challenging and controversial topics, due to the poor survival rate and potentially devastating effects on speech and swallowing [1]. Alcohol consumption and acetaldehyde, a toxic product of ethanol metabolism, are widely known as carcinogen of head and neck SCC (HNSCC) and esophageal SCC (ESCC). The activity of aldehyde dehydrogenase 2, a key enzyme in the elimination of aldehyde, is reduced by the germline polymorphism Glu504Lys (previously described as Glu487Lys), which is prevalent in Mongoloid but not in Caucasoid or Negroid populations [2]. Therefore, this different genetic background is considered as a major reason of high HPSCC and ESCC incidence rates in East Asia [3, 4].
Tumor suppressor gene TP53 is the most frequently mutated gene in human cancers: more than 50% of human cancers contain somatic mutations in this gene [5, 6]. Tumor suppressor p53, encoded by the TP53 gene, is a key protein involved in many cellular anticarcinogenic processes such as apoptosis and cell-cycle control [7]; therefore, p53 is widely known as the guardian of the genome [8]. Molecular alterations in carcinogenesis of HNSCC include loss of p53 function, which is mediated by genetic mechanisms such as TP53 mutations [9] and loss of heterozygosity [10], or degradation of p53 meditated by the human papillomavirus (HPV) oncoprotein E6 [11].
Two studies previously demonstrated the association between TP53 mutations and prognosis in surgically treated HNSCC patients. [12, 13] However, these studies did not examine these associations based on the anatomical location of the HNSCCs. Moreover, patients with oropharyngeal SCC (OPSCC) comprised the majority of the cases, and there were a total of only two patients with HPSCC in the two studies. HPV-related OPSCCs commonly express wild type TP53 [14], creating a potential confounder as HPV-related tumors have a generally favorable prognosis. In contrast, HPV-driven HPSCC is considered rare [15] and the prognostic significance of TP53 mutation status in HPSCC has not yet been investigated. The aim of this study was to evaluate the prognostic significance of TP53 mutation status among surgically treated HPSCC patients in Japan, where the HPSCC incidence rate is high.
Methods
We retrospectively reviewed the clinical charts of HPSCC patients, who had been surgically treated between 2008 and 2014 at the University of Tokyo Hospital. We excluded patients, who underwent salvage surgery after the definitive radiotherapy (RT) or chemoradiotherapy (CRT), and those who received preoperative chemotherapy. We identified 57 HPSCC patients (55 men and 2 women; age range: 46–84 years, median age: 68 years) who underwent initial surgery of primary lesions. Subsites of primary tumor were the pyriform sinus, posterior wall, and postcricoid region, in 37 (65%), 15 (26%), and 5 (9%) patients, respectively. TNM staging was done according to the 7th edition of the Union for International Cancer Control (2009) staging guidelines. The indication for postoperative RT/CRT was comprehensively determined on the basis of the clinicopathological status of the patients including impaired performance status, inadequate surgical margin, ≥4 metastatic LNs, presence of extranodal extension (ENE), and postoperative complications as well as the consent of patient. The Institutional Review Board of the University of Tokyo Hospital approved this study (#2487 and #2904).
Determination of human papillomavirus status
In OPSCC, p16 immunopositivity is commonly used as a surrogate marker for HPV determination [16]. Therefore, the p16 status was evaluated in surgically excised specimens using immunohistochemistry (IHC) according to the standard techniques as previously described [17]. A mouse p16 monoclonal antibody (1:100 dilution; Santa Cruz Biotechnology, CA, USA) was used as the primary antibody, and immunostained samples were blindly reviewed and scored independently by two investigators (M. A and Y. S). In accordance with previous studies, p16 positivity by IHC was defined as strong and diffuse nuclear and cytoplasmic staining in ≥70% of the tumor cells [16, 17].
However, p16 expression does not always indicate the presence of HPV DNA, and the combination of p16 expression determined by IHC with HPV DNA determination by polymerase chain reaction (PCR) or in situ hybridization (ISH) is considered to provide the almost perfect sensitivity and specificity [18, 19]. Therefore, p16-immunopositive specimens were also tested for HPV DNA by HPV-ISH, as previously described [19, 20]. Briefly, HPV DNA was detected using an ISH method with catalyzed signal amplification (GenPoint signal amplification system; Dako, Kyoto, Japan), in accordance with the manufacturer’s instructions. Slides were hybridized using a biotinylated GenPoint HPV probe (This probe has been found to react with HPV types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 68 on FFPE tissues and/or cells by ISH, Dako). Slides were scored as positive for HPV if a punctate signal pattern was observed in almost all tumor nuclei.
Genomic DNA extraction
Tumor tissue specimens were collected during surgery, and snap-frozen in liquid nitrogen and stored at −80 °C. Genomic DNA was extracted using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany), in accordance with the manufacturer’s protocol. In specimens where the harvest of frozen sections appeared to interfere with the pathological margins, DNA was isolated from formalin-fixed, paraffin-embedded (FFPE) tissue blocks. Briefly, the tumor lesions on hematoxylin and eosin-stained slides were marked, and the corresponding areas were identified on unstained tissue sections. Each selected area was carefully dissected under microscopic observation. Genomic DNA was then extracted using the QIAamp DNA FFPE Tissue Kit (Qiagen).
Detection of TP53 mutations
PCR amplification and Sanger sequencing were performed to detect TP53 mutations in exons 2–9, containing 98% of all mutations described in HNSCC cases [21]. A total of 20 ng/μl genomic DNA per sample was used for PCR amplification using PrimeSTAR HS DNA Polymerase(Takara Bio, Shiga, Japan). Amplification conditions included two-step cycle of 98 °C for 15 s and 68 °C for 90 s, for a total of 44 cycles, except for the amplification of exon 2–3 fragments harvested from frozen and FFPE specimens and exon 6 fragments harvested from FFPE specimens, which were amplified by nested PCR (25 cycles each) using two primer pairs. Subsequently, PCR products harvested from FFPE tissue were purified using the QIAquick PCR Purification Kit (Qiagen), in accordance with the manufacturer’s protocol. Mutations were confirmed by Sanger sequencing using the Big Dye Terminator v3.1 Cycle Sequencing Kit and 3130xL Genetic Analyzer (Applied Biosystems, CA, USA). In this study, nonsense mutations, splice variants, and frameshifts were defined as truncating mutations, that lead to nonfunctional p53, based on previous studies [13, 22]. All samples were sequenced twice with independent PCR using forward and reverse primers.
Immunohistochemistry for p53 expression
IHC for p53 expression was performed according to standard IHC techniques. A mouse p53 monoclonal antibody clone DO-7 (1:100 dilution; Leica Biosystems, Nussloch, Germany) was used as the primary antibody. In accordance with a previous study, a sample was determined as p53-immunopositive when ≥10% of tumor nuclei were immunostained [23].
Statistical analyses
Primary endpoint was disease-specific survival (DSS) and secondary endpoint was overall survival (OS). Potential correlations between the treatment method and several clinical features were evaluated using the chi-square test; for analyses in which there were <4 patients, the Fisher’s exact test was used. Survival was analyzed using the Kaplan–Meier method and the log-rank test. Variables were also analyzed by multivariate survival analysis using the Cox proportional hazards model. Hazard ratios (HR) and 95% confidence intervals (CI) were calculated to determine the effect of each variable on outcomes. P values <0.05 were considered statistically significant. GraphPad Prism software version 5 (GraphPad Software, CA, USA) was used for the chi-square, Fisher’s exact, Mann-Whitney’s U, and log-rank tests. Mac Tahenryo-Kaiseki version 2.0 (ESUMI, Tokyo, Japan) was used for multivariate Cox regression models.
Results
Human papillomavirus status of patients with hypopharyngeal squamous cell carcinoma
In this cohort of 57 HPSCC patients, 3 (5%) patients were immunopositive for p16; however, none of these three patients had detectable HPV DNA by HPV-ISH. Therefore, HPSCC was confirmed to be unrelated to HPV in all patients in this study (Fig. 1).
Distribution of TP53 mutations
TP53 mutations were detected in 39 (68%) patients. Missense mutations, nonsense mutations, splicing variants, and frameshift mutations were found in 24 (42%), 9 (16%), 4 (7%), and 2 (3%) patients, respectively. TP53 mutations in exon 2, 3, 4, 5, 6, 7, 8, and 9 were found in 1, 0, 3, 11, 9, 4, 8, and 3 patients, respectively (Fig. 2).
Clinicopathological features and TP53 mutation status
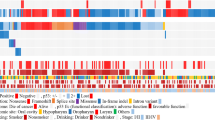
Table 1 summarizes the clinical data and TP53 status. Table 2 shows the clinicopathological features according to TP53 mutation status. Histopathological analysis revealed positive surgical margins in 11 (19%) patients, ≥4 metastatic LNs in 14 (25%) patients, and ENE in 15 (26%) patients. Postoperative RT/CRT was administered to 16 (28%) patients. Of note, all of stage I/II patients (5 patients) had wild-type TP53, and patients with a past history of HNSCC or ESCC were significantly greater in the TP53 mutation groups than in the wild-type groups (P = 0.02). Administration of postoperative RT/CRT did not correlate with TP53 mutation status (P = 0.25).
Association between TP53 mutation and p53 expression
Representative images of specimens exhibiting p53 immunopositivity are presented in Fig. 3. p53 immunopositivity was detected in 22 patients, including 5 (28%) and 17 (71%) patients in the wild-type and missense mutation groups, whereas there were no patients with p53 immunopositivity in the truncating mutation group (P = 0.0001, chi-square test).
Correlation between TP53 mutation status and prognosis
Eighteen (32%) patients died from HPSCC, whereas 8 (14%) patients died from other causes. The remaining 31 (54%) patients were alive and disease-free on last follow-up date. The median follow-up period for the entire cohort was 29 months (range: 3.5–101 months), whereas 45 months (range: 24–101 months) for patients who survived (n = 31) and 16 months (range: 3.5–85 months) for those who died (n = 26). The 3-year DSS of the wild-type group was significantly longer than that of the TP53 mutation group (94% vs 55%; P = 0.01, Fig. 4). Furthermore, patients with wild-type/missense mutations had significantly better 3-year DSS than those with truncating mutations (76% vs 43%; P = 0.03). The 3-year DSS rate of wild-type, missense mutation, and truncating mutation groups were 94%, 61%, and 43%, respectively (Fig. 5). The 3-year OS of the wild-type group was significantly longer than that of the TP53 mutation group (89% vs 42%; P = 0.007). The 3-year OS rate of wild-type/missense mutation group was not significantly different than that of the truncating mutation group (66% vs 40%; P = 0.14). The 3-year OS rate of the wild-type, missense mutation, and truncating mutation group were 89%, 43%, and 40%, respectively. In contrast, p53 immunopositivity did not correlate with DSS (P = 0.77). In the subgroup analyses of 52 stage III/IV patients, the 3-year DSS of the wild-type group was significantly longer than that of the TP53 mutation group (92% vs 55%; P = 0.02). The 3-year OS of the wild-type group was significantly longer than that of the TP53 mutation group (92% vs 42%; P = 0.006).
Table 3 shows the associations between the clinicopathological factors and DSS in univariate analysis. The presence of ≥4 metastatic LNs (P = 0.04) and ENE (P = 0.03) were poor prognostic factors. In contrast, tumor differentiation grade, T classification, stage, surgical margin, and postoperative RT/CRT did not correlate with DSS.
Multivariate Cox proportional hazard analysis using variables based on univariate analyses was conducted to determine independent prognostic factors for DSS and OS. The presence of TP53 mutations (P = 0.04; HR, 4.96; 95% CI, 1.08–22.8, and P = 0.02; HR, 4.75; 95% CI, 1.35–16.7, respectively) and ≥4 metastatic LNs (P = 0.03; HR, 3.00; 95% CI, 1.12–8.04, and P = 0.02; HR, 2.89; 95% CI, 1.22–6.86, respectively) have significant adverse effects on both DSS and OS. In the subgroup analyses of 52 stage III/IV patients, the presence of TP53 mutations was a significant adverse prognostic factor on OS, and nearly reached significance on DSS. (Table 4).
Discussion
In this retrospective study, we demonstrated that the TP53 mutation status significantly correlated with poor prognosis in surgically treated HPSCC patients. Specifically, patients with truncating mutations exhibited the worst prognosis. To the best of our knowledge, this is the first study focusing on the association between TP53 mutation status and prognosis of HPV-unrelated HPSCC. The result of the current study was consistent with the previous studies investigating all HNSCC subsites [12, 13], which included patients with HPV-driven OPSCC.
HPSCC is rarely caused by HPV-driven carcinogenesis as we confirmed in the current study and occurs more frequently in East Asian population than in other regions of the world. Survival of patients with HPSCC has not markedly improved in recent decades. In the last two-decades, CRT and induction chemotherapy followed by RT have become the option for advanced HNSCC patients who prefer nonsurgical organ preservation [24,25,26]. However, RT-induced late toxicity, such as dysphasia and osteonecrosis distresses emerging issues for cancer survivors. The recent development of minimally invasive surgical procedures, such as transoral robotic surgery (TORS) and transoral videolaryngoscopic surgery (TOVS) techniques, has broadened surgical indications and appeared to result in better outcomes with respect to the postoperative speech and swallowing function [27, 28]. Therefore, surgery remains the main treatment modality for HPSCC patients. Our multivariate analyses revealed that both the TP53 mutation status and the presence of ≥4 metastatic LNs were independent adverse prognostic factors for surgically treated HPSCC patients. In the previous study, we demonstrated that the presence of multiple metastatic LNs was significantly associated with the poor prognosis and the incidence of distant metastases in advanced HPSCC patients treated with total pharyngolaryngectomy [29]. Collectively, TP53 mutations can be a useful biomarker for HPSCC patients, in addition to the traditional metastatic LN number.
Interestingly, the past history of HNSCC or ESCC was significantly higher in the TP53 mutation group than in the wild-type group in the present study. HNSCC and ESCC have been known to occur synchronously or metachronously, which might be explained by the concept of “field cancerization” first introduced by Slaughter et al. in 1953 [30]. Currently, repetitive exposure to acetaldehyde is considered to play a key role in field cancerization of the squamous epithelium in the head and neck region and the esophagus [31]. Moreover, Waridel et al. reported that mutations in TP53 were frequent and early events in the pathogenesis of HNSCC and identified the expansion of multiple clones of mutant p53-containing cells as an important biological step in field cancerization [32]. Our findings in the current study led further support to these observations. Future studies with larger sample size and longitudinal evaluations, supported with basic research, are necessary to confirm this hypothesis.
In the current study, we demonstrated that p53 immunopositivity was observed most frequently in the presence of missense mutations. Wild-type p53 protein is rapidly degraded via the ubiquitin-proteasome system, resulting in low p53 protein expression. Conversely, the nonsense-mediated RNA decay and the resultant decreased amount of the protein considered to be the reason why truncating p53 proteins cannot be detected by IHC [33]. Some missense mutations, that result in increased p53 immunopositivity can lead to a dominant-negative or a gain-of-function phenotype [34, 35], Our observations in the current study support these biological mechanisms; therefore, the distinction between missense and truncating mutations is reasonable for the clinical categorization of the TP53 mutation status.
The Cancer Genome Atlas (TCGA) reported that TP53 mutations were detected in 84% of HPV-unrelated HNSCC cases using whole-exome sequencing analysis [14]. In comparison, the frequency of TP53 mutations was lower in the HPSCC cohort of the current study, which might be partially due to differences in racial composition and tumor subsites. The HNSCC cohort of TCGA consisted almost entirely of Caucasoid and Negroid populations, with only two HPSCC patients. Additionally, it is possible that mutation detection sensitivity of whole-exome sequencing was superior to that of Sanger sequencing.
To improve the prognoses of HPSCC patients with TP53 mutations, adjuvant therapy should be selectively administered to these patients. TP53 mutation, however, is also known as a predictive marker for chemo- and radioresistance in HNSCCs. [36, 37] Therefore, it might be unreasonable to use TP53 mutation status as a therapeutic biomarker for existing postoperative treatments including RT/CRT. Although most of the current targeted therapies are inhibitors of oncogenic pathways, development of p53-targeted therapy is warranted.
One of the limitations of our study was a lack of detailed comparison and functional study of each TP53 mutations, due to the small sample size. In line with previous reports on HNSCC [13], various mutation types were detected in various regions of TP53 genes. Further investigation with larger sample size is required to elucidate the potential associations between the mutations with respect to functional and biological effects and prognosis. Furthermore, the number of T1–2 tumors was small in this study. Recently, TORS and TOVS techniques for T1–2 tumors to which RT/CRT was previously preferable were broadened. Therefore, further accumulation of T1–2 patients is also required.
Conclusions
We demonstrated that TP53 mutations had a significant impact on prognosis, in surgically treated HPSCC patients. In particular, truncating mutations which were not detected by p53 IHC were shown to have predictive value for a worst survival. Further confirmation from prospective studies with larger sample size including more T1–2 patients is warranted.
Abbreviations
- CRT:
-
Chemoradiotherapy
- ENE:
-
Extranodal extension
- ESCC:
-
Esophageal squamous cell carcinoma
- FFPE:
-
Formalin-fixed, paraffin-embedded
- HNSCC:
-
Head and neck squamous cell carcinoma
- HPSCC:
-
Hypopharyngeal squamous cell carcinoma
- HPV:
-
Human papillomavirus
- IHC:
-
Immunohistochemistry
- ISH:
-
In situ hybridization
- LN:
-
Lymph node
- ND:
-
Neck dissection
- OPSCC:
-
Oropharyngeal squamous cell carcinoma
- PCR:
-
Polymerase chain reaction
- RT:
-
Radiotherapy
- SCC:
-
Squamous cell carcinoma
- TCGA:
-
The Cancer Genome Atlas
- TORS:
-
Transoral robotic surgery
- TOVS:
-
Transoral videolaryngoscopic surgery
References
Montgomery PQ, Phys Evan PH, Gullane PJ. Principles and practice of head and neck surgery and oncology. 2nd ed. Informa Healthcare: Colchester; 2009. p. 233.
Goedde HW, Agarwal DP, Fritze G, Meier-Tackmann D, Singh S, Beckmann G, et al. Distribution of ADH2 and ALDH2 genotypes in different populations. Hum Genet. 1992;88:344–6.
Hamajima N, Takezaki T, Tajima K. Allele frequencies of 25 polymorphisms pertaining to cancer risk for Japanese, Koreans and Chinese. Asian Pac J Cancer Prev. 2002;3:197–206.
Asakage T, Yokoyama A, Haneda T, Yamazaki M, Muto M, Yokoyama T, et al. Genetic polymorphisms of alcohol and aldehyde dehydrogenases, and drinking, smoking and diet in Japanese men with oral and pharyngeal squamous cell carcinoma. Carcinogenesis. 2007;28:865–74.
Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–31.
Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23.
Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–10.
Efeyan A, Serrano M. p53: guardian of the genome and policeman of the oncogenes. Cell Cycle. 2007;6:1006–10.
Olshan AF, Weissler MC, Pei H, Conway K. p53 mutations in head and neck cancer: new data and evaluation of mutational spectra. Cancer Epidemiol Biomark Prev. 1997;6:499–504.
Gonzalez MV, Pello MF, Lopez-Larrea C, Suarez C, Menendez MJ, Coto E. Loss of heterozygosity and mutation analysis of the p16 (9p21) and p53 (17p13) genes in squamous cell carcinoma of the head and neck. Clin Cancer Res. 1995;1:1043–9.
Scheffner M, Werness BA, Huibregtse JM, Levine AJ, Howley PM. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–36.
Poeta ML, Manola J, Goldwasser MA, Forastiere A, Benoit N, Califano JA, et al. TP53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357:2552–61.
Lindenbergh-van der Plas M, Brakenhoff RH, Kuik DJ, Buijze M, Bloemena E, Snijders PJ, et al. Prognostic significance of truncating TP53 mutations in head and neck squamous cell carcinoma. Clin Cancer Res. 2011;17:3733–41.
The Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517:576–82.
Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tân PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363:24–35.
Gillison ML, Zhang Q, Jordan R, Xiao W, Westra WH, Trotti A, et al. Tobacco smoking and increased risk of death and progression for patients with p16-positive and p16-negative oropharyngeal cancer. J Clin Oncol. 2012;30:2102–11.
Saito Y, Yoshida M, Ushiku T, Omura G, Ebihara Y, Shimono T, et al. Prognostic value of p16 expression and alcohol consumption in Japanese patients with oropharyngeal squamous cell carcinoma. Cancer. 2013;119:2005–11.
Thavaraj S, Stokes A, Guerra E, Bible J, Halligan E, Long A, et al. Evaluation of human papillomavirus testing for squamous cell carcinoma of the tonsil in clinical practice. J Clin Pathol. 2011;64:308–12.
Saito Y, Yoshida M, Omura G, Kobayashi K, Fujimoto C, Ando M, et al. Prognostic value of p16 expression irrespective of human papillomavirus status in patients with oropharyngeal carcinoma. Jpn J Clin Oncol. 2015;45:828–36.
Saito Y, Ebihara Y, Ushiku T, Omura G, Kobayashi K, Ando M, et al. Negative human papillomavirus status and excessive alcohol consumption are significant risk factors for second primary malignancies in Japanese patients with oropharyngeal carcinoma. Jpn J Clin Oncol. 2014;44:564–9.
Bouaoun L, Sonkin D, Ardin M, Hollstein M, Byrnes G, Zavadil J, Olivier M. TP53 Variations in Human Cancers: New Lessons from the IARC TP53 Database and Genomics Data. Hum Mutat. 2016;37(9):865–76.
Oliver M, Langer A, Carrieri P, Bergh J, Klaar S, Eyfjord J, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–67.
Rodrigo JP, Martínez P, Allonca E, Alonso-Durán L, Suárez C, Astudillo A, et al. Immunohistochemical markers of distant metastasis in laryngeal and hypopharyngeal squamous cell carcinomas. Clin Exp Metastasis. 2014;31:317–25.
Pignon JP, Bourhis J, Domenge C, Designe L. Chemotherapy added to locoregional treatment for head and neck squamous-cell carcinoma: three meta-analyses of updated individual data. MACH-NC collaborative group. Meta-analysis of chemotherapy on head and neck cancer. Lancet. 2000;355:949–55.
Posner MR, Hershock DM, Blajman CR, Mickiewicz E, Winquist E, Gorbounova V, et al. Cisplatin and fluorouracil alone or with docetaxel in head and neck cancer. N Engl J Med. 2007;357:1705–15.
Posner MR, Norris CM, Wirth LJ, Shin DM, Cullen KJ, Winquist EW, et al. Sequential therapy for the locally advanced larynx and hypopharynx cancer subgroup in TAX 324: survival, surgery, and organ preservation. Ann Oncol. 2009;20:921–7.
Park YM, Kim YS, De Virgilio A, Lee SY, Seol JH, Kim SH. Transoral robotic surgery for hypopharyngeal squamous cell carcinoma: 3-year oncologic and functional analysis. Oral Oncol. 2012;48:560–6.
Tomifuji M, Araki K, Yamashita T, Shiotani A. Transoral videolaryngscopic surgery for oropharyngeal, hypopharyngeal, and supraglottic cancer. Eur Arch Otorhinolaryngol. 2014;271:589–97.
Omura G, Ando M, Saito Y, Kobayashi K, Yamasoba T, Asakage T. Disease control and clinicopathological prongostic factors of total pharyngolaryngectomy for hypopharyngeal cancer: a single-center study. Int J Clin Oncol. 2015;20:290–7.
Slaughter DP, Southwick HW, Smejkal W. Field cancerization in oral stratified squamous epithelium; clinical implications of multicentric origin. Cancer. 1953;6:963–8.
Ohashi S, Miyamoto S, Kikuchi O, Goto T, Amanuma Y, Muto M. Recent advances from basic and clinical studies of esophageal squamous cell carcinoma. Gastroenterology. 2015;149:1700–15.
Waridel F, Estreicher A, Bron L, Flaman JM, Fontolliet C, Monnier P, et al. Field cancerisation and polyclonal p53 mutation in the upper aero-digestive tract. Oncogene. 1997;14:163–9.
Ebihara Y, Iwai M, Akashi K, Ito T, Omura G, Saito Y, et al. High incidence of null-type mutations of the TP53 gene in Japanese patients with head and neck squamous cell carcinoma. J Cancer Ther. 2014;5:664–71.
Petitjean A, Achatz MIW, Borresen-Dale AL, Hainaut P, Olivier M. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene. 2007;26:2157–65.
Xu Y. Induction of genetic instability by gain-of-function p53 cancer mutants. Oncogene. 2008;27:3501–7.
Perrone F, Bossi P, Cortelazzi B, Locati L, Quattrone P, Pierotti MA, et al. TP53 mutations and pathologic complete response to neoadjuvant cisplatin and fluorouracil chemotherapy in resected oral cavity squamous cell carcinoma. J Clin Oncol. 2010;28:761–6.
Skinner HD, Sandulache VC, Ow TJ, Meyn RE, Yordy JS, Beadle BM, et al. TP53 disruptive mutations lead to head and neck cancer treatment failure through inhibition of radiation-induced senescence. Clin Cancer Res. 2012;18:290–300.
Acknowledgements
We thank Ms. A. Tsuyuzaki for her excellent technical assistance.
Funding
This study was supported by JSPS KAKENHI Grant Number 15 K20184 and 26,893,058.
Availability of data and materials
All datasets supporting the conclusions of this article are included in Table 1. Primer sequences for TP53 sequencing are available upon request.
Author information
Authors and Affiliations
Contributions
Conceptualization: GO, MA, Methodology: GO, TA, Validation: MA, YS, Formal analysis: GO, YS, Investigation: GO, MA, YE, Resources: KK, OF, KA, MY, Data curation: OF, Writing – original draft: GO, Writing – review and editing: MA, Visualization: GO, Supervision: TA, TY, Project administration: MA, TA, Funding acquisition: GO, MA. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The Institutional Review Board of the University of Tokyo Hospital approved this study (#2487 and #2904). Written informed consent was obtained from all the patients.
Consent for publication
Written informed consent was obtained from all the patients.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Omura, G., Ando, M., Ebihara, Y. et al. The prognostic value of TP53 mutations in hypopharyngeal squamous cell carcinoma. BMC Cancer 17, 898 (2017). https://doi.org/10.1186/s12885-017-3913-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-017-3913-1