Abstract
Background
Although eradication therapy for chronic Helicobacter pylori (H. pylori) reduces the risk of gastric cancer (GC), its effectiveness is not complete. Therefore, it is also critically important to identifying those patients who remain at high risk after H. pylori eradication therapy. Accumulation of protein methylation is strongly implicated in cancer, and recent study showed that dimethylation of eEF1A lysine 55 (eEF1AK55me2) promotes carcinogenesis in vivo. We aimed to investigate the relationship between eEF1A dimethylation and H. pylori status, efficacy of eradication therapy, and GC risk in H. pylori-eradicated mucosa, and to reveal the potential downstream molecules of eEF1A dimethylation.
Methods
Records of 115 patients (11 H. pylori-negative, 29 H. pylori-positive, 75 post-eradication patients) who underwent upper gastrointestinal endoscopy were retrospectively reviewed. The eEF1A dimethyl level was evaluated in each functional cell type of gastric mucosa by immunofluorescent staining. We also investigated the relationship between eEF1AK55me2 downregulation by CRISPR/Cas9 mediated deletion of Mettl13, which is known as a dimethyltransferase of eEF1AK55me2.
Results
The level of eEF1A dimethylation significantly increased in the surface and basal areas of H. pylori-positive mucosa compared with the negative mucosa (surface, p = 0.0031; basal, p = 0.0036, respectively). The eEF1A dimethyl-levels in the surface area were significantly reduced by eradication therapy (p = 0.005), but those in the basal area were maintained even after eradication therapy. Multivariate analysis revealed that high dimethylation of eEF1A in the basal area of the mucosa was the independent factor related to GC incidence (odds ratio = 3.6611, 95% confidence interval = 1.0350–12.949, p = 0.0441). We also showed the relationship between eEF1A dimethylation and expressions of reprogramming factors, Oct4 and Nanog, by immunohistochemistry and in vitro genome editing experiments.
Conclusions
The results indicated that H. pylori infection induced eEF1A dimethylation in gastric mucosa. The accumulation of dimethyl-eEF1A in the basal area of the mucosa might contribute to GC risk via regulation of reprograming factors in H. pylori eradicated-gastric mucosa.
Similar content being viewed by others
Background
Helicobacter pylori (H. pylori) infection, which causes atrophic gastritis, has been defined as a sequence of histological events that confers an increasing risk of malignant transformation [1, 2]. Indeed, previous basic studies have provided evidence that infection with H. pylori carrying specific virulence factors can lead to gastric carcinogenesis [3, 4]. Although, the success rate of eradication therapy has fallen, due to rising resistance to clarithromycin [5], the provision of eradication for H. pylori infection by national health insurance was started in February 2013 to reduce the number of new cases of gastric cancer (GC) in Japan.
Reductions in primary and metachronous GC are expected with H. pylori eradication, but eradication alone does not completely eliminate GC risk in patients with atrophic gastric mucosa as precancerous lesions [6]. Even after eradication therapy and follow-up diagnosis with barium and endoscopy, there is an urgent need to improve an effective follow-up strategy to identify patients at high risk for GC.
In previous studies, accumulations of methylation in gastric mucosa after H. pylori eradication therapy were correlated with GC risk [7]. Most of these reports pointed out only epigenetic biomarkers, such as DNA or histone methylation, in GC risk related to chronic gastritis [8]. However, it was recently revealed that hundreds and likely thousands of proteins are also methylated at lysine, which leads to activation of cellular pathways, such as those for growth signaling and response to damage from chronic inflammation [9]. Eukaryotic elongation factor 1A (eEF1A) is one of the evolutionarily conserved and fundamental non-ribosomal components of the translational machinery [10]. The expression or upregulation of eEF1A has been reported to be associated with cancer development and invasion in ovarian, breast, lung, prostate, hepatic, and pancreatic cancers [11,12,13,14]. Additionally, a recent study showed that dimethylation of eEF1Al lysine 55 (eEF1AK55me2) leads to its activation and utilization to increase translational output and promote carcinogenesis in vivo [15].
Although methylation is recognized as a key mechanism in cancer development in atrophic gastric mucosa [16, 17], how lysin methylations are maintained in atrophic gastric mucosa and contribute to GC development is mostly unclear. In this study, we hypothesized that H. pylori infection would contributes to potential GC risk through the dimethylation of eEF1A, an active GTPase form, in gastric mucosa.
The aim of this study was thus to clarify the association between the dimethylation of eEF1A, H. pylori infection status, and GC incidence in gastric mucosa. Additionally. We investigated how dimethylated eEF1A contributes to GC development after H. pylori eradication in gastric mucosa by focusing on the reprogramming factors Oct4 (octamer-binding transcription factor 4) and Nanog.
Materials and methods
Study subjects and sampling
We retrospectively investigated the mucosa of the middle portion of the greater curvature of the gastric corpus, approximately 8 cm from the gastric cardia, in patients who underwent upper gastrointestinal endoscopy with biopsy at Oita University Hospital, Japan, between January 2001 and December 2018. Of the 150 enrolled patients, 27 patients with unknow eradication time and 8 patients after gastrectomy were excluded from this study; the remaining 115 patients were included in this study (Additional file 1: Fig. S1). H. pylori infection was determined by culture, histology, and rapid urease test. In H. pylori-negative patients, we additionally confirmed serum antibody titer (< 3 U/mL) and the lack of endoscopic atrophy and histologic gastritis (Additional file 1: Fig. S1). The success of H. pylori eradication therapy was defined as a negative 13C-urease breath test that was confirmed within 6 months after the therapy. Additionally, to exclude H. pylori-reinfected patients, we confirmed absence of H. pylori by culture, rapid urease test, and histology in eradicated gastric mucosa.
The biopsy site “B2”, which is recommended by the Updated Sydney System, is assumed to be suitable for evaluating the histological status of fundic glands [18]. Specimens were collected endoscopically from the greater curvature of the gastric middle bodies. Biopsy specimens were immediately fixed in 10% neutral buffered formalin for 24 h and embedded in paraffin wax blocks. Fixed samples were sliced into 3-μm-thick sections and stained with hematoxylin and eosin (HE). Degrees of inflammation, activity, atrophy, and intestinal metaplasia were scored according to the Updated Sydney System (0, none; 1, weak; 2, moderate; 3, marked) [18].
Immunofluorescent staining
Anti-MUC5AC (1:50, Leica Biosystems), H+/K+ATPase (1:100; Medical & Biological Laboratories), MUC6 (1:200; BioRad), and PG1 (1:200; Abnova) antibodies were used for immunostaining to differentiate between cell types in the fundic gland. We performed immunohistochemistry using anti-dimethyl-eEF1AK55me2 antibody (1:400; Abclonal), Nanog (1:10,000; Cell Signaling Technology), and Oct4 (1:100; Santa Cruz Biotechnology). All biopsy materials were fixed in 10% buffered formalin for 24 h and then embedded in paraffin. After deparaffinization and subsequent rehydration to remove xylene, endogenous peroxidases were inactivated with 3% hydrogen peroxide solution (Wako, Osaka, Japan). Antigen retrieval was performed at pH 6.0. Next, the sections were incubated overnight with monoclonal anti-MUC5AC, MUC6, H+/K+ATPase, PG1, and dimethyl-eEF1A primary antibodies, followed by incubation with secondary antibodies (1:1000; Alexa Fluor R488 and 594) at room temperature for 2 h. After washing, images were captured with a Keyence BZ-9000 microscope (Keyence, Osaka, Japan).
Evaluation of dimethyl eEF1A level
Dimethylation levels of eEF1A were evaluated on surface, middle and basal areas of gastric glands, which were identified by morphological appearance or immunostaining with antibodies for specific markers, MUC5AC (surface area), H + /K + ATPase (middle area), and PG1 (basal area), respectively. After immunofluorescence staining with anti-dimethyl-eEF1A antibody, mean fluorescent intensities on these areas were quantified using the regions of interest (ROI) tool in ImageJ software. Multiple ROIs were set for each area based on marker immunofluorescence in one image, and several images at 4 × magnification were used for each patient. Then, the dimethylation level of eEF1A for each area in each patient was expressed as an average of multiple mean fluorescent intensities from the ROIs.
Cell culture and CRISPR/Cas9
TMK-1, a gastric cell line, was provided by Dr. H. Ito (Tottori Prefectural Kousei Hospital, Tottori, Japan) with permission from an original provider, Dr. A. Ochiai (National Cancer Center, Kashiwa, Japan). Cells were cultured in RPMI1640 (Sigma) with 10% fetal bovine serum. We established Mettl13, which is the physiologic eEF1A lysine 55 dimethyltransferase [15], knockout TMK-1cells using the CRISPR-Cas9 genome editing system and generated stable Mkettl13 knockout TMK-1cells. Briefly, a single guide (sgRNA) targeting Mettl13 (5′-AAGAAAGCTTTCGAGTGGTATGG-3′) was cloned into a Cas9-expressing plasmid (pSpCas9(BB)-2A-Puro(PX450); Addgene plasmid #62,988), and the plasmid was trandfected into TMK-1 cells, which was subsequently cloned after a 2-day treatment with 2 µg/mL puromycine. Knockout of Mettl13 was confirmed by Western blotting.
Western blotting
Western blotting was performed as described previously [19]. Cells were lysed on ice for 20 min in SDS-modified RIPA buffer containing protease and phosphatase inhibitor cocktails (cOmplete Mini; Roche Diagnostics, Mannheim, Germany) (PhosSTOP EASYpack; Roche Diagnostics), and then centrifuged at 4 °C at 15,000 rpm for 20 min. The resulting cell lysates (20 µg each) were boiled with Laemmli sample buffer and subjected to SDS-PAGE. The samples were transferred to a polyvinylidene difluoride membrane (Merk Millipore, Darmstadt, Germany), which was blocked for 1 h in Block Ace (DS Pharma, Osaka, Japan) at room temperature, then incubated overnight at 4 °C with primary antibodies against METTL13 (1:1000; Bethyl Laboratories), dimethyl-eEF1A (1:1000; ABclonal Technology), eEF1A(1:1000; Abclonal technology), Nanog(1:1000; Abcom), Oct4(1:1000, Abcom), and GAPDH (1:1000, Santa Cruz Biotechnology, Santa Cruz). After washing with 1 × TBS containing 0.1% Tween 20, the membranes were incubated for 1 h at room temperature with appropriate secondary antibodies [19], followed by rewashing. Finally, the signals were detected using an ECL Western blotting analysis system (GE Healthcare, Piscataway, NJ, USA) in accordance with the manufacturer’s instructions.
Statistical analysis
We investigated the relationship between the clinical factors and dimethyl-eEF1A level of atrophic gastritis mucosa using univariate and multivariate analysis. All variables are expressed as mean ± standard deviation for continuous data (Table 2). Prior to analysis, continuous data were divided into two groups using average values. Levels of eEF1A dimethylation were compared between groups using analysis of variance or analysis of covariance (for age-adjusted data), followed by the Tukey test. Univariate analyses were performed using the Student t-test or the Tukey test for continuous variables and chi-squared test for categorical variables. The results of the multivariate logistic regression analysis are expressed as adjusted odds ratios with 95% confidence intervals. A p-value < 0.05 was considered to indicate statistical significance. All statistical analyses were performed using with JMP® 11 (SAS Institute Inc, Cary, NC, USA).
Results
We retrospectively investigated the gastric corpus mucosa from 115 subjects (11 H. pylori-negative, 29 H. pylori-positive, 75 post-eradication patients). The characteristic of the patients are provided in Table 1. Updated Sydney System scores for inflammation, activity, atrophy, and intestinal metaplasia (IM) are shown in Table 2. The dimethyl level of eEF1A of each gland area identified morphologically or by immunostaining for specific markers (surface, MUC5A; middle, H + /K + ATPase; and basal, PG1) was evaluated (Additional file 2: Fig. S2, Table 2). In the middle area, dimethyl-eEF1A was not detected in MUC6-positive cells but only in H + /K + ATPase-positive cells (Additional file 2: Fig. S2).
Difference in eEF1A dimethylation of gastric mucosa by H. pylori infection status
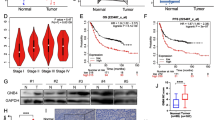
To investigate the association between H. pylori infection/eradication and eEF1A dimethylation in gastric mucosa, we first compared the dimethylation levels of eEF1A in each area (MUC5AC-, H+/K+ATPase-, and PG1-positive cells) among the three groups, H. pylori-negative, H. pylori- positive, and post-eradicated patients (Fig. 1A). H. pylori-positive patients showed higher eEF1A dimethylation than H. pylori-negative patients in the surface and basal areas of gastric mucosa (surface: 16.91 ± 6.42 vs. 28.11 ± 10.96, p = 0.0004; basal: 14.03 ± 4.88 vs. 29.23 ± 6.33, p < 0.0001, respectively). These results suggested that dimethylation of eEF1A is associated with H. pylori infection in gastric mucosa.
Immunofluorescent staining of dimethyl-eEF1A in H. pylori-negative,-positive, and -eradicated gastric mucosa. A Representative images of immunofluorescent staining using an antibody against level of dimethyl-eEF1A (eEF1AK55me2). Surface, middle, and basal areas were defined by co-staining with markers specific for each area (see Additional file 1: Fig. S1). B Levels of dimethyl-eEF1A in surface, middle, and basal areas were compared between H. pylori-negative, -positive, and post-eradicated patients. Differences were calculated by Mann–Whitney U test. *p < 0.05; **p < 0.01; ***p < 0.001
The dimethylated eEF1A that was caused by H. pylori infection was effectively reduced in the surface area (MUC-5AC-positive area) of post-eradicated mucosa (p = 0.0027) (Fig. 1B). However, H. pylori eradication therapy did not affect eEF1A dimethylation in the basal area (PG-1-positive area) (Fig. 1B). These results suggested that the dimethylation of eEF1A in the basal level remained high in atrophic gastric mucosa even after H. pylori eradication therapy.
Univariate and multivariate analyses of GC and eEF1A dimethylation in basal area of gastric mucosa
Next, we investigated whether dimethylation of eEF1A in the basal area could cause a risk of cancer in post-eradicated atrophic gastritis mucosa. The influence of known potential risk factors for GC (age, sex, period after eradication therapy, and histological features) and eEF1A dimethyl level in basal mucosa were analyzed in 75 H. pylori eradication patients, which included in part those with a history of GC treatment. Univariate analyses showed that severe histological atrophy and high eEF1A dimethylation of the basal mucosal area were the risk factors for GC (Fig. 2, Table 3). Multivariate analysis revealed that not only histological atrophic levels (odds ratio = 9.9493, 95% confidence interval = 1.7096–57.901, p = 0.0106) but also high dimethylation of eEF1A in basal mucosal areas (odds ratio = 3.6611, 95% confidence interval = 1.0350–12.949, p = 0.0441) were the independent factors related to GC incidence (Table 3).
Immunofluorescent staining of dimethyl-eEF1A in H. pylori-eradicated mucosa with or without GC incidence. A Representative images of immunofluorescent staining using an antibody against level of dimethyl-eEF1A (eEF1AK55me2) and PG-1 for H. pylori-eradicated gastric mucosa. B Levels of dimethyl-eEF1A in basal areas were compared between post-eradicated patients with or without GC incidence. Differences were calculated by Mann–Whitney U test. *p < 0.05; **p < 0.01; ***p < 0.001
Accumulation of eEF1A dimethylation and metaplasia in gastric mucosa
Metaplastic lineages have been associated with various injurious scenarios in atrophic gastritis mucosa. We investigated eEF1A dimethylation in the IM area (n = 11) of eradicated mucosa. There was no significant accumulation in eEF1A dimethylation in the IM area compared with other gastric gland cells (mean: 22.01 ± 8.86, Table 2). Although IM is considered an important step in the pathogenesis of GC in atrophic gastric mucosa, there was no significant increase in eEF1A dimethylation in the IM area in eradicated patients with GC incidence (GC(-), n = 7 and GC( +), n = 4) (Fig. 3).
Comparison of dimethyl-eEF1A in area of intestinal metaplasia with or without GC incidence. A Representative images of immunofluorescent staining using an antibody against level of dimethyl-eEF1A (eEF1AK55me2) for eradicated gastric mucosa. B Levels of dimethyl-eEF1A were compared between patients with or without a incidence of GC using Mann–Whitney U test
Association of eEF1A dimethylation with the expressions of reprogramming factors Oct4 and Nanog
Next, we investigated whether the expressions of Oct4 and Nanog correlate with dimethylated eEF1A in post-eradicated mucosa. As shown in Fig. 4A, B, the proportion of Oct4- or Nanog-positive cells in eradicated mucosa was significantly higher in patients with high eEF1A dimethylation. However, there was no significant increase in Oct4- or Nanog-positive cells in the IM area (Additional file 3: Fig. S3). To investigate whether dimethylated eEF1A contributes to the expressions of Oct4 and Nanog in gastric cells, we next deleted Mettl13 in a TMK-1 cell using the CRISPR/Cas9 system (Fig. 4C) and analyzed the effect on the expressions of Oct4 and Nanog. As shown in Fig. 4D and Additional file 4: Fig. S4, knockout of Mettl13 led to a decrease of the eEF1A dimethylation level in TMK-1 cells. Interestingly, reduction of dimethylated eEF1A downregulated the expression of Oct4 and Nanog in TMK-1 cells (Fig. 4E). These results suggest that accumulation of dimethylated eEF1A in the basal area of eradicated mucosa is associated with the expression of Oct4 and Nanog, which control reprogramming of various types of differentiated cells, in response to chronic damage induced by H. pylori infection.
Levels of eEF1A dimethylation were compared with expressions of Oct4 and Nanog in H. pylori-eradicated tissues and a Mettl13-deleted cell line. A Representative images of immunofluorescent staining using antibodies against dimethyl-eEF1A, Oct4 and Nanog in H. pylori-eradicated mucosa. B The numbers of Oct4- or Nanog-positive cells per gland in H. pylori-eradicated mucosa were compared between patients with low levels (Oct4: n = 7, Nanog: n = 8) and those with high levels (Oct4: n = 7, Nanog: n = 8) of eEF1A dimethylation by Mann–Whitney U test. C and D Knockout of Mettl13 and downregulation of eEF1A dimethylation in TMK-1 cell were confirmed by Western blotting. E Western blotting with antibodies against Oct4, Nanog, and GAPDH revealed that knockout of Mettl13 in TMK-1 cell caused downregulation of Oct4 and Nanog expressions
Discussion
The presence of H. pylori and virulence factors such as CagA, a well-known inducer of chronic inflammation and atrophy, have been reported to be associated with aberrant DNA methylation in gastric mucosa [17, 20, 21]. However, it remains unknown whether H. pylori infection also induces aberrant methylation at the protein level, except for histone modification. In this study, we immunohistochemically analyzed the association of H. pylori status with the distribution of dimethylated eEF1A, whose function in tumorigenicity has been characterized recently by Liu S et al. [15]. This is first report, to our knowledge, to spatially show the impact of H. pylori infection on the level of eEF1A dimethylation in gastric mucosa.
Our results showed that the levels of eEF1A dimethylation in gastric mucosa were different according to the H. pylori status (negative, positive, and post-eradication therapy). We found that eEF1A dimethylation of surface and basal cell areas was higher in H. pylori-positive patients than in -negative patients, suggesting that H. pylori infection induce eEF1A dimethylation of surface and basal cells in gastric mucosa. Indeed, eEF1A dimethylation in H. pylori-negative mucosa with adenocarcinoma of the fundic gland type, which is known to be unrelated to H. pylori infection, was as low as those in H. pylori-negative and cancer-free mucosa (data not shown), supporting our hypothesis that H. pylori infection might cause aberrant protein methylation.
Furthermore, not only inflammation but also aging is known to induce accumulation of methylation in multi-organ tissues. Because there were age differences between these three groups (Table 1), we also calculated the age-adjusted levels of eEF1A dimethylation and compared them between these groups using analysis of covariance with the Tukey test. We found the same tendency in both the crude and the age-adjusted models, indicating that age-related differences in methylation did not account for the results (Fig. 1B).
Eradication of H. pylori infection has been reported to reduce the risk of GC among asymptomatic patients in high-risk countries [6]. However, long-term studies from Japan revealed that eradication therapy cannot reduce the risk of GC completely [22,23,24]. In this study, we focused on a subset of atrophic gastritis patients who showed high dimethylation of eEF1A in the basal area of H. pylori-eradicated mucosa and found that the high dimethylation of eEF1A in PG1-positive cells, which are chief cells in the basal area of gastric mucosa, correlated with GC incidence in post-eradicated gastric mucosa (Additional file 6: Table S1). Unlike that in the basal area of H. pylori-eradicated gastric mucosa, eEF1A dimethylation in the surface area (MUC5AC-positive cells) was strongly associated with the H. pylori eradication therapy, i.e., that in the surface area was significantly reduced after eradication regardless of the GC incidence. Considering that the cell lineages in gastric mucosa are known to vary greatly in life span: from 3 to 5 days for surface cells to several months for basal chief cells [25], these different life spans among gastric-cell lineages might cause the specific accumulation of eEF1A dimethylation in the basal area even after eradication therapy. In our results, eEF1A dimethylation in the basal area of eradicated mucosa did not show significant reduction even after the long-term monitoring after H. pylori eradication therapy in patients with GC incidence (follow up periods of < 3.5 vs. ≥ 3.5 years, p = 0.3908). Although data from further long-term monitoring after eradication therapy is needed, the level of dimethylated eEF1A could be a potential marker for GC risk during the follow up of H. pylori eradicated patients. We additionally evaluated the level of dimethylated eEF1A in gastric mucosa using biopsy samples by Western blotting (data not shown). Although dimethyl-eEF1A levels were detectable, they were not appropriate for a cell-by-cell analysis. Further study will be required to use the dimethyl-eEF1A level as a minimally invasive biomarker for the prediction of GC risk in eradicated mucosa.
Although the severity of atrophic damage is already known to be strongly associated with carcinogenesis by inducing various oncogenic alterations, such as overexpression or amplifications, in gastric mucosa [26, 27], the pathogenic mechanism of how H. pylori infection induces GC has been a challenging question for decades. Consistent with the previous studies, the present study also showed that the patients with severe atrophic change have GC incidence (Table 3). Additionally, we confirmed that eEF1A dimethylation in the basal area of gastric mucosa is also an independent factor of GC incidence. Previous studies have also shown that damage in the gastric mucosa, including H. pylori-induced oxyntic atrophy, initiates mucosal metaplasia and the reprogramming and proliferation of basal cells, and these response are associated with carcinogenesis in atrophic gastric mucosa [28,29,30]. Although in our results, there was no significant increase in dimethylated eEF1A in the metaplasia area in eradicated gastric mucosa with GC incidence, we showed the association between the expressions of Oct4 and Nanog, which are known as crucial reprogramming factors, and eEF1A dimethylation in the basal area of eradicated gastric mucosa. Additionally, we also showed that dimethyl-eEF1A induced the expression of Oct4 and Nanog in TMK-1 cells. In recent reports, the chief cell has been identified as a cell capable of reprogramming and a metaplastic response to parietal cell damage in atrophic gastric mucosa [28, 31]. Oct4 and Nanog lie in the core of the transcriptional network that controls stem cell pluripotency [32]. Self-renewing progenitor and differentiating cells expressing reprograming factors may be the target for cancer initiation and progression, through the induction of symmetrical cell division [27]. Although their roles in atrophic tissue are largely unknown, these results suggests that high eEF1A dimethylation in the basal area of eradicated mucosa not only could be a useful marker for GC risk, but also might cooperatively contribute to the development of gastric lesions, such as metaplasia and cancer, via regulation of Oct4 and Nanog. Further molecular functional analysis of eEF1A dimethylation, such as METTL13 rescue experiments for METTL13-KO TMK-1 cells, would lead to elucidation of the mechanism of carcinogenesis in H. pylori eradicated-gastric mucosa.
The present study has several limitations. First, this was a retrospective observational study performed at a single center. It only included patients who underwent upper gastrointestinal endoscopy for abdominal symptoms or other medical problems reported. Most participants had primary disease; therefore, bias may have occurred. Next, our study contained only 11 patients with IM, 4 of whom had a history of GC treatment (Table 3). In our study, dimethyl-eEF1A was not detected in MUC6-positive cells (Additional file 5: Fig. S5), so we therefore focused only on the fundic gland area. Although there was no significant difference in eEF1A dimethylation in the IM area between patients with or without GC incidence, some IM regions showed a high level of eEF1A dimethylation even after eradication therapy. Because a recent study showed that alterations in methylation were reported to have been observed in regions of IM and to be associated with subsequent dysplasia or GC in gastric mucosa [33], further prospective studies with larger numbers of samples including IM regions in the antrum area of the gastric mucosa may lead us to the discovery of new pathological mechanisms in carcinogenesis in eradicated gastric mucosa.
In conclusion, the results indicated that H. pylori infection induces dimethylation of eEF1A in the surface and basal areas of gastric mucosa. Especially, the dimethylation level of eEF1A in the basal area of gastric mucosae may be associated with the expression of reprogramming factors and GC risk in post-H. pylori-eradicated mucosa.
Availability of data and materials
Not applicable.
References
Amieva M, Peek RM Jr. Pathobiology of Helicobacter pylori-Induced gastric cancer. Gastroenterology. 2016;150(1):64–78.
Malfertheiner P, Megraud F, O’Morain CA, Atherton J, Axon AT, Bazzoli F, Gensini GF, Gisbert JP, Graham DY, Rokkas T, et al. Management of Helicobacter pylori infection–the Maastricht IV/ Florence consensus report. Gut. 2012;61(5):646–64.
Chang WL, Yeh YC, Sheu BS. The impacts of H. pylori virulence factors on the development of gastroduodenal diseases. J Biomed Sci. 2018;25(1):68.
Díaz P, Valenzuela Valderrama M, Bravo J, Quest AFG. Helicobacter pylori and gastric cancer: adaptive cellular mechanisms involved in disease progression. Front Microbiol. 2018;9:5.
Sue S, Kuwashima H, Iwata Y, Oka H, Arima I, Fukuchi T, Sanga K, Inokuchi Y, Ishii Y, Kanno M, et al. The superiority of vonoprazan-based first-line triple therapy with clarithromycin: a prospective multi-center cohort study on Helicobacter pylori eradication. Intern Med (Tokyo, Japan). 2017;56(11):1277–85.
Lee YC, Chiang TH, Chou CK, Tu YK, Liao WC, Wu MS, Graham DY. Association between helicobacter pylori eradication and gastric cancer incidence: a systematic review and meta-analysis. Gastroenterology. 2016;150(5):1113–24.
Usui G, Matsusaka K, Mano Y, Urabe M, Funata S, Fukayama M, Ushiku T, Kaneda A. DNA methylation and genetic aberrations in gastric cancer. Digestion. 2021;102(1):25–32.
Wang H, Li NS, He C, Xie C, Zhu Y, Lu NH, Hu Y. Discovery and validation of novel methylation markers in Helicobacter pylori-associated gastric cancer. Dis Markers. 2021;2021:4391133.
Biggar KK, Li SSC. Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol. 2015;16(1):5–17.
Negrutskii BS, El’skaya AV. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol. 1998;60:47–78.
Tomlinson VA, Newbery HJ, Wray NR, Jackson J, Larionov A, Miller WR, Dixon JM, Abbott CM. Translation elongation factor eEF1A2 is a potential oncoprotein that is overexpressed in two-thirds of breast tumours. BMC Cancer. 2005;5:113.
Li R, Wang H, Bekele BN, Yin Z, Caraway NP, Katz RL, Stass SA, Jiang F. Identification of putative oncogenes in lung adenocarcinoma by a comprehensive functional genomic approach. Oncogene. 2006;25(18):2628–35.
Yang S, Lu M, Chen Y, Meng D, Sun R, Yun D, Zhao Z, Lu D, Li Y. Overexpression of eukaryotic elongation factor 1 alpha-2 is associated with poorer prognosis in patients with gastric cancer. J Cancer Res Clin Oncol. 2015;141(7):1265–75.
Duanmin H, Chao X, Qi Z. eEF1A2 protein expression correlates with lymph node metastasis and decreased survival in pancreatic ductal adenocarcinoma. Hepatogastroenterology. 2013;60(124):870–5.
Liu S, Hausmann S, Carlson SM, Fuentes ME, Francis JW, Pillai R, Lofgren SM, Hulea L, Tandoc K, Lu J, et al. METTL13 methylation of eEF1A Increases translational output to promote tumorigenesis. Cell. 2019;176(3):491–504.
Asada K, Nakajima T, Shimazu T, Yamamichi N, Maekita T, Yokoi C, Oda I, Ando T, Yoshida T, Nanjo S, et al. Demonstration of the usefulness of epigenetic cancer risk prediction by a multicentre prospective cohort study. Gut. 2015;64(3):388–96.
Maekita T, Nakazawa K, Mihara M, Nakajima T, Yanaoka K, Iguchi M, Arii K, Kaneda A, Tsukamoto T, Tatematsu M, et al. High levels of aberrant DNA methylation in Helicobacter pylori-infected gastric mucosae and its possible association with gastric cancer risk. Clin Cancer Res. 2006;12(3 Pt 1):989–95.
Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney system. International workshop on the histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20(10):1161–81.
Hirashita Y, Tsukamoto Y, Kudo Y, Kakisako D, Kurogi S, Hijiya N, Nakada C, Uchida T, Hirashita T, Hiratsuka T, et al. Early response in phosphorylation of ribosomal protein S6 is associated with sensitivity to trametinib in colorectal cancer cells. Lab Invest. 2021;101(8):1036–47.
Roesler BM, Rabelo-Gonçalves EM, Zeitune JM. Virulence factors of Helicobacter pylori: a review. Clin Med Insights Gastroenterol. 2014;7:9–17.
Muhammad JS, Eladl MA, Khoder G. Helicobacter pylori-induced DNA methylation as an epigenetic modulator of gastric cancer: recent outcomes and future direction. Pathogens. 2019;13;8(1):23.
Michigami Y, Watari J, Ito C, Nakai K, Yamasaki T, Kondo T, Kono T, Tozawa K, Tomita T, Oshima T, et al. Long-term effects of H. pylori eradication on epigenetic alterations related to gastric carcinogenesis. Sci Rep. 2018;8(1):14369.
Take S, Mizuno M, Ishiki K, Yoshida T, Ohara N, Yokota K, Oguma K, Okada H, Yamamoto K. The long-term risk of gastric cancer after the successful eradication of Helicobacter pylori. J Gastroenterol. 2011;46(3):318–24.
Murakami K, Kodama M, Nakagawa Y, Mizukami K, Okimoto T, Fujioka T. Long-term monitoring of gastric atrophy and intestinal metaplasia after Helicobacter pylori eradication. Clin J Gastroenterol. 2012;5(4):247–50.
Mills JC, Shivdasani RA. Gastric epithelial stem cells. Gastroenterology. 2011;140(2):412–24.
Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P, et al. Kyoto global consensus report on Helicobacter pylori gastritis. Gut. 2015;64(9):1353–67.
Deng N, Goh LK, Wang H, Das K, Tao J, Tan IB, Zhang S, Lee M, Wu J, Lim KH, et al. A comprehensive survey of genomic alterations in gastric cancer reveals systematic patterns of molecular exclusivity and co-occurrence among distinct therapeutic targets. Gut. 2012;61(5):673–84.
Nam KT, Lee HJ, Sousa JF, Weis VG, O’Neal RL, Finke PE, Romero-Gallo J, Shi G, Mills JC, Peek RM Jr, et al. Mature chief cells are cryptic progenitors for metaplasia in the stomach. Gastroenterology. 2010;139(6):2028–37.
Willet SG, Lewis MA, Miao ZF, Liu D, Radyk MD, Cunningham RL, Burclaff J, Sibbel G, Lo HG, Blanc V, et al. Regenerative proliferation of differentiated cells by mTORC1-dependent paligenosis. EMBO J. 2018;3;37(7):e98311.
Al-Marzoqee FY, Khoder G, Al-Awadhi H, John R, Beg A, Vincze A, Branicki F, Karam SM. Upregulation and inhibition of the nuclear translocation of Oct4 during multistep gastric carcinogenesis. Int J Oncol. 2012;41(5):1733–43.
Burclaff J, Willet SG, Sáenz JB, Mills JC. Proliferation and differentiation of gastric mucous neck and chief cells during homeostasis and injury-induced metaplasia. Gastroenterology. 2020;158(3):598-609.e595.
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122(6):947–56.
Huang KK, Ramnarayanan K, Zhu F, Srivastava S, Xu C, Tan ALK, Lee M, Tay S, Das K, Xing M, et al. Genomic and epigenomic profiling of high-risk intestinal metaplasia reveals molecular determinants of progression to gastric cancer. Cancer Cell. 2018;33(1):137–50.
Acknowledgements
We would like to thank Ms. Kanako Ito, and Ms. Yoko Kudo for their excellent assistance with the experiments.
Author information
Authors and Affiliations
Contributions
All authors have approved the final version of the article. Conceptualization: YH; Formal analysis and investigation: YH, MF, KK, TF and MK; Writing—original draft preparation: YH; Writing—review and editing: KMizukami, YT, KH, TO, MK; Resources: SO, YK, YW, KT, KF, KO, RO, and OM; Supervision: KMurakami. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethic approval and consent to participate
This study was approved by the ethics committee of Oita University Faculty of Medicine (approval number: 1541). All procedures were carried out according to the principles outlined in the Declaration of Helsinki.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Fig. S1
Flow chart of patients enrolled in this study
Additional file 2: Fig. S2
This study profile and immunofluorescent staining of H. pylori-eradicated gastric mucosa. A All patients received eradication therapy and were enrolled at least 6 months after establishment of successful H. pylori eradication (n=75). B Schematic model of a fundic gland. C Representative images of immunofluorescent staining for MUC5AC, MUC6, H+/K+ATPase, PG1, and dimethyl-eEF1A of gastric mucosa are shown. Merging was not detected in the MUC6 and dimethyl-eEF1A cells
Additional file 3: Fig. S3
Representative images of immunofluorescent staining using antibodies against dimethyl-eEF1A, Oct4, and Nanog in IM area
Additional file 4: Fig. S4
Image of full-length membranes of Western blotting
Additional file 5: Fig. S5
Levels of dimethyl-eEF1A in MUC6-positive areas were compared between H. pylori-negative, -positive, and post-eradicated patients. Differences were calculated by Mann–Whitney U test. *p <0.05; **p <0.01; ***p <0.001
Additional file 6: Table S1
. Raw data of fluorescence intensity of dimethyl-eEF1A
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hirashita, Y., Fukuda, M., Kodama, M. et al. Potential association of eEF1A dimethylation at lysine 55 in the basal area of Helicobacter pylori-eradicated gastric mucosa with the risk of gastric cancer: a retrospective observational study. BMC Gastroenterol 22, 490 (2022). https://doi.org/10.1186/s12876-022-02521-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-022-02521-5