Abstract
Background
This study aimed to identify the transcription factors of nitrate reductase genes (NIA1 and NIA2) promoters and hypothetical cis-element of NRE2. Based on the constructed cDNA library of Nicotiana tabacum K326, a yeast one-hybrid system was established using the Matchmaker® Gold Yeast One-Hybrid Library Screening System from Clontech. The transcription factors of NIA1 andNIA2 promoters and NRE2 cis-elements were screened.
Results
After sequencing and bioinformatics analysis, 15 cDNA sequences were identified: 9 for NIA1 (including XP_016503563.1 and NP_001312236.1), 3 for NIA2 (including XP_016510250.1), and 3 for NRE2 (including XM_016576899.1). XP_016503563.1 was annotated in PREDICTED: CRM-domain containing factor CFM3, and NP_001312236.1chloroplastic/mitochondrial-like in Nicotiana tabacum. NP_001312236.1 was annotated in Sulfite oxidase-like of Nicotiana tabacum. XP_016510250.1 was annotated as PREDICTED: uncharacterized protein LOC107827596 in Nicotiana tabacum. XM_016576899.1 was annotated in PREDICTED: Nicotiana tabacum RING-H2 finger protein ATL16-like (LOC107759033).
Conclusion
A yeast one-hybrid library was successfully constructed. The identified transcription factors may provide a theoretical basis for the study of plant nitrate reductase.
Similar content being viewed by others
Background
Nitrogen is a fundamental constituent of cell components such as nucleic acids, amino acids, proteins, membranes, and cell walls. Additionally, nitrogen is quantitatively the most important nutrient that plants acquire from soil [1]. Nitrogen deficiency exerts severe pressure on plant growth, development, and photosynthesis, ultimately limiting plant productivity [2,3,4]. Nitrate is a primary nitrogen source for most plants [5]. Furthermore, it functions as a signal molecule that promptly triggers changes in metabolism, growth, and gene expression in plants [6, 7]. The genes encoding the enzymes of nitrate reductase (NR) and nitrite reductase (NIR) are typical nitrate-inducible genes [8]. Overexpression of NR and NIR genes has an influence on the nitrogen metabolism of plants. Davenport et al. [9] reported that overexpression of the NIR gene in tobacco could affect both nitrogen and carbon metabolism, extend stay-green time, elevate NR activity, reduce nitrate, proline, glutamine, and glutamate contents.
Plant growth, development, and their responses to the environment are regulated by the differential expression of genes, of which the major mechanism is transcriptional regulation controlled by transcription factor (TF) which binds to DNA cis-elements located in the gene promoters [10]. Currently, several identified TFs and other factors that regulate transcription are involved in nitrate signal transduction, such as MADS-box TF Arabidopsis nitrate-regulated 1 (ANR1) [11], NIN-like protein 7 (NLP7) [12], and the lateral organ boundary domain (LBD) gene family transcription factors of LBD37, LBD38, and LBD39 [13]. Furthermore, previous studies have shown the presence of a conserved sequence (GACCCTT-N (9–10)-AAG), a nitrate-responsive cis-element (NRE), located in the NIR gene promoter in a variety of plants that is necessary for gene expression regulation in the nitrate-mediated signal transduction pathway [14, 15]. Nevertheless, until now, the TF acting on the NR gene promoter and NRE that responds to a nitrate signal in the promoter remains unclear.
According to the existing NRE sequence, we found that there were similar but slightly different sequences (GACCCTA-N (10)-AAG) in the NR genes (NIA1 and NIA2) promoters of Nicotiana tabacum, which was tentatively named as NRE2. Our present study aimed to investigate the role of NRE2 in NIA1 and NIA2 promoters and to identify the TFs specifically binding to NIA1, NIA2, and NRE2 based on a yeast one-hybrid system. The findings of this study may not only help to further understand the regulatory pathway of nitrate signal response in tobacco but also provide new ideas for future research into nitrate signal transduction in other plants.
Methods
Cloning of NIA1and NIA2 promoters
We searched the ATG upstream sequences of NIA1and NIA2genes in the National Center of Biotechnology Information (NCBI) database, and a 937 bp sequence was intercepted from each gene for the design of primers. The primers were as follows: NIA1 (forward), TATATATATGACCCTGCAATGAAAG; NIA1 (reverse), AGATTATTCTAAAAAAGAAAGAGAGAT; NIA2 (forward), TACATACAAGGGCGCGAATAA; and NIA2 (reverse), AGATTATTCTAAAAAAAGAATATGAATG. Using the tobacco genome as template, polymerase chain reaction (PCR) amplification was performed to generate fragments which were then sequenced and compared with the target fragments.
Construction of the β-glucuronidase (GUS) expression vector
The tobacco cultivar “Nicotiana benthamiana” was from the national tobacco cultivation physiological and biochemical base, Agricultural University Of He’nan, China. And Agrobacterium (EHA105; Shanghai Zeye biotechnology co., LTD, China) was used for was used for the transformation experiments. The possible NO3− signal response element (NRE2) sequence (GACCCTACGGGCGTAAAAAG) was predicted from PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/), a database of plant cis-acting regulatory elements, enhancers, and repressors [16]. After the analysis of promoter sequences of NIA1 and NIA2 genes, we found NRE2 is located at -281 bp ~ − 262 bp upstream of NIA1 gene and -332 bp ~ − 313 bp upstream of NIA2 gene. Next, the NIA1 and NIA2 promoters were modified by the knockout of NRE2 element, and inserting four tandem NRE2 elements. The primers used for amplification of modified promoters were the same as the primers described above. The 35S promoter of vector pCAMBIA-NPT-GUS was digested with EcoR I and Hind III. The original and modified NIA1 and NIA2 promoters were digested with EcoR I and Hind III, respectively, and inserted in place of the 35S sequence in pCAMBIA-NPT-GUS using T4 DNA ligase. The ligated products were then transformed into Escherichia coli DH5α competent cells and recombinant clones were identified using colony PCR. Recombinant expression vectors that expressed GUS genes [pCAMBIA-NPT-NIA1-P-GUS, pCAMBIA-NPT-NIA1-P-△NRE2-GUS (meaning without the NRE2 element), pCAMBIA-NPT-NIA1-P-(4 × NRE2)-GUS (meaning with four tandem NRE2 elements), pCAMBIA-NPT-NIA2-P-GUS, pCAMBIA-NPT-NIA2-P-△NRE2-GUS, and pCAMBIA-NPT-NIA2-P-(4 × NRE2)-GUS] were selected. The recombinant plasmids were validated by PCR.
Agrobacterium-mediated transient transfection assay
Nicotiana benthamiana (grown for 45 days) was used in this experiment. Agrobacterium EHA105 was grown overnight in 5 mL YEB medium supplemented with kanamycin (50 μg/mL), rifampicin (25 μg/mL), and streptomycin (50 μg/mL). Next, 1.5 mL bacterial solution was transferred into a sterile tube and centrifuged for 10 min at 1000×g. After washing, the bacterial solution was diluted 10-fold to obtain an optical density (OD) of 0.3–0.4 at 600 nm. The tobacco plant was placed under a white fluorescent lamp for 1 h to ensure that the stomata were fully opened. Two large leaves were selected and the cuticle was removed through friction (about 0.5 cm2). The Agrobacterium solution was injected into the area lacking a cuticle using a 1-mL syringe. After 48 h of transfection, the leaf tissue was collected and assayed immediately.
Histochemical GUS staining and quantitative measurements
GUS reporter gene enzymatic activity was determined using histochemical staining. Briefly, the mature leaves of transformants were collected and cut into small round pieces using a punch. These small round pieces were stained with GUS staining solution for 6 h at 37 °C, followed by elution with 50, 70, and 100% ethanol until completely decolorized. Additionally, GUS activity was quantitative analyzed using a fluorometric assay, with 4-umbelliferyl-D-glucuronide as substrate according to a previous study [17]. Protein content was determined via bradford protein assay [18].
Acquisition of bait sequence
The upstream promoter sequences (about 1 kb) of NIA1 and NIA2 were cloned based on primers. To improve the screening efficiency, four NRE2 sequences were tandemly iterated. After ligating Xho I and Hind III sites that were the same in all vector plasmids to both ends of the sequences above, bait sequences with four tandem NRE2 elements (4 × NRE2) were obtained and named “4 box.”
Construction of a tobacco cDNA library
Tobacco (K326), provided by the tobacco cultivation and physiology laboratory of Henan Agricultural University, was used in this experiment. A cDNA library of K326 was constructed via a switching mechanism at the 5′ end of RNA transcript (SMART™). Briefly, the root, stem, leaf, and flower of K326 at different developmental stages were quickly frozen in liquid nitrogen and then stored at − 70 °C. Total RNA from different tissues was extracted and mixed together. After purification of total RNA using NucleoSpin RNA II (740,955.20; Clontech, Palo Alto, CA, USA), poly A+ mRNA was enriched using a NucleoTrap mRNA Kit (740,655; Clontech). The first strand of cDNA was then synthesized based on the SMART technique and duplex-specific nuclease (DSN) according to the Matchmaker® Gold Yeast One-Hybrid Library Screening System (Clontech). Double-stranded cDNA was synthesized through long-distance (LD)-PCR. Following purification using a chromatographic column, a normalized cDNA library of K326 was obtained.
Construction of a recombinant bait plasmid
The NIA1 and NIA2 promoter sequences, 4 box oligonucleotides, and the pAbAi vector were digested with XhoI and HindIII, respectively. The enzyme-digested products were purified on agarose gel and ligated into the linearized bait sequences and pAbAi vector (carrying the URA3and AUR1-C genes) using T4 ligase. The ligated products were transformed into E. coli DH5α competent cells and positive recombinant clones were screened using Luria-Bertani solid media containing ampicillin. The plasmids were then extracted and digested with Xho I and Hind III before PCR electrophoresis to confirm the insertion of the target fragment. The recombinant plasmids were identified by DNA sequencing.
Transformation of linearized bait plasmids into yeast cells
Yeast competent cells were prepared using the LiAc method. BbsB I-linearized recombinant bait plasmids (pAbAi-P: NIA1, pAbAi-P: NIA2, and pAbAi-P: (4 × NRE2)) (1 μg) were integrated into the genome of yeast (Y1HGold). The transformed competent cells were then transferred onto solid agar synthetic defined (SD) medium-Ura and incubated for 3 days. Single clones were then identified via colony PCR using Matchmaker Insert Check PCR Mix 1 (Clontech). The expected size of PCR products was 1.35 kb.
Determination of basal expression of the reporter gene
Healthy yeast colonies grown in SD medium-Ura were selected and resuspended in 0.9% NaCl. After adjusting to obtain OD600nm values of approximately 0.002, yeast strain was transferred onto each of the following solid media: SD-Ura without aureobasidin A (AbA), SD-Ura with 100 ng/mL AbA, SD-Ura with 200 ng/mL AbA, SD-Ura with 300 ng/mL AbA, SD-Ura with 400 ng/mL AbA, SD-Ura with 500 ng/mL AbA, SD-Ura with 600 ng/mL AbA, SD-Ura with 700 ng/mL AbA, SD-Ura with 800 ng/mL AbA, SD-Ura with 900 ng/mL AbA, and SD-Ura with 1000 ng/mL AbA. The colonies were allowed to grow for 2–3 days at 30 °C. The minimum AbA concentration that completely inhibited colony growth was determined and used for further library screening.
Construction and screening of a yeast one-hybrid library
Purified double-stranded cDNA (5 μg) and SmaI-linearized pGADT7-Recexpression vector (3 μg) were mixed to co-transform bait yeast using the Yeast maker Yeast Transformation System 2 (Clontech). Yeast cell suspensions were diluted to 1/10, 1/100, 1/1000, and 1/10000, and 100-μL diluents were coated onto SD-Leu plates to screen the total number of colonies. The remaining yeast cell suspensions (approximately 15 mL) were coated on corresponding SD-Leu/AbA plates (200 μL per plate) to preliminarily screen the positive colonies. The number of colonies on SD-Leu was calculated 3–5 days later using the following formula: total number of colonies = colony-forming unit/volume of coated plate × dilution rate × total volume of cell suspension.
Confirmation of positive interaction and plasmid extraction
Healthy single clones from the SD-Leu/AbA plate were transferred into new SD-Leu medium containing AbA and incubated at 30 °C for 3–5 days. The screened yeast single clones were then transferred onto SD-Leu/AbA for repeated screening. The colonies that eventually grew on SD-Leu/AbA plates were selected for yeast colony PCR using Matchmaker Insert Check PCR Mix 2 (Clontech) to obtain the inserted fragment of cDNA library. A small amount of the product was identified by 1% tris-acetate-ethylenediamine tetraacetic acid (TAE) agarose/ethidium bromide (EtBr) gel electrophoresis. After purification, the PCR products were sequenced at the Beijing Genomics Institute, Shenzhen, China, with T7 as primer. Bioinformatic analyses were then performed on the data to analyze the cDNA library quality and identify positive clones. Sequences with biological significance were selected and the corresponding plasmids were extracted using an Easy Yeast Plasmid Isolation Kit (630,467; Clontech) and stored at − 20 °C.
Statistical analysis
All data processing was done using SPSS 21.0 software, and the measurement data have been expressed in mean ± SD. Statistical differences between two groups were tested by Student’s t-test., and a P < 0.05 indicates statistical significance.
Results
Cloning of NIA1 and NIA2 promoters
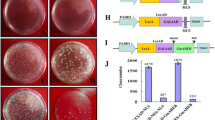
The tobacco NIA1 and NIA2 gene promoter sequences were cloned based on the designed primers. The amplified products were analyzed by agarose gel electrophoresis, and their lengths were consistent with target lengths (Fig. 1a). Sequencing results, as shown in Fig. 1b, suggest that NIA1 and NIA2 promoter sequences were successfully cloned.
Cloning of NIA1 and NIA2 promoters and GUS expression levels. Electrophoretic diagram of cloned NIA1 promoter and NIA2 promoter (a). M: DNA marker; P-NIA1: the amplified fragment of NIA1 gene promoter; P-NIA2: the amplified fragment of the NIA2 gene promoter. Sequencing results of cloned (i) NIA1 promoter and (ii) NIA2 promoter (b). GUS histochemical staining results of tobacco leaves transformed by different promoter sequences (c) and the GUS content of tobacco leaves transformed by different promoter sequences measured by quantitative assay (d). * represents P < 0.05, and ** represents P < 0.01 when compared with normal NIA1 promoters; # represents P < 0.05 when compared with normal NIA2 promoters
GUS activity assay
In order verify whether the change of NRE2 elements could influence the expression of NIA1 and NIA2 genes, we added four NRE2 in NIA1 and NIA2 promoters ((pCAMBIA-NPT-NIA1-P-(4 × NRE2)-GUS and pCAMBIA-NPT-NIA2-P-(4 × NRE2)-GUS)), which will lead to the improvement of the screening efficiency. Moreover, the GUS expression can be directly reflected by staining. As shown in Fig. 3c, the results of GUS staining showed that, when compared with the control (staining results of the leaves transformed by normal NIA1 and NIA2 promoters), GUS expression levels significantly decreased after knockout of the NRE2 elements in NIA1 and NIA2 promoters. Additionally, GUS expression levels in the leaves transformed by NIA1 and NIA2 promoters containing 4 tandem NRE2 elements were significantly higher than that in the leaves transformed by NIA1 and NIA2 promoters lacking NRE2 sequences. Moreover, a quantitative assay also revealed that knockout of the NRE2 sequences in the NIA1 and NIA2 promoters could significantly reducethe GUS expression levels compared to those of the control, respectively (both P < 0.05; Fig. 1d). Moreover, the leaves transformed by NIA2 promoter containing 4 tandem NRE2 elements showed higher GUS expression (more than 10%) than leaves in the control group (both, P < 0.05).
Identification of bait yeast strain and determination of AbA basal expression
Three bait recombinant vectors (pAbAi-P: NIA1, pAbAi-P:NIA2, and pAbAi-P: (4 × NRE2)) were transformed into Y1HGold. Colony PCR showed that the obtained fragment length was consistent with the expected size of PCR products (1.35 kb not including the inserted fragment, Fig. 2a), suggesting that the three bait recombinant vectors were successfully transformed into yeast cells.
Identification of bait-yeast strains by yeast colony PCR (a). NIA1p: PCR product of colony transformed by pAbAi-NIA1p plasmid; NIA2p: PCR product of colony transformed by pAbAi-NIA2p plasmid; 4box: PCR product of colony transformed by pAbAi-(4 × NRE2) plasmid; Marker: DL2000 DNA marker. Identification of the minimum inhibitory concentration of AbA (b)
To omit the influence of target sequence recognition by endogenous yeast transcription factors, the minimum inhibitory concentration of AbA (basal expression level of reporter gene) was measured. As shown in Fig. 2b, the minimum concentrations of AbA needed to suppress basal expression of the NIA1, NIA2, and 4box bait strains were 400 ng/mL, 800 ng/mL, and 800 ng/mL, respectively.
Construction of a K326 cDNA library
Electrophoretic identification of the constructed K326 cDNA library showed that the bands were dispersive and evenly distributed (Fig. 3a), indicating that the expression of each gene in the library was average. This result suggests that the K326 normalized cDNA library was successfully constructed.
Electrophoretic diagram of cDNA library (a). M: DNA marker; cDNA: electrophoretic feature of constructed cDNA library. Colony map of calculating library capacity (b). i, Growth of 1/100 dilution of NIA1P transformant; ii, Growth of 1/100 dilution of NIA2P transformant; iii: Growth of 1/100 dilution of 4 × NRE2 transformant
Screening of a yeast one-hybrid library and extraction of yeast plasmids
NIA1P, NIA2P, and 4box yeast cell suspensions were diluted to 1/100, and 304, 264, and 240 colonies, respectively, were obtained on SD-Leu plates (Fig. 3b). The total number of colonies was 4.6 × 106, 3.96 × 106, and 3.6 × 106, respectively. Colony PCR and electrophoretic identification showed that the insert fragment was 200 bp to 2000 bp in length, indicating that the capacity and quality of the constructed library was adequate.
The identification analysis with PCR assay was performed on a total of 310 positive colonies with NIA1 promotor sequences, and 97 PCR products with bright bands and long fragments were sequenced and analyzed via bioinformatics. Finally, 9 sequences of Nicotiana were selected, including 7 predicted proteins (such as XP_016503563.1), 1 hypothetical protein, and 1 sulfite oxidase-like protein (NP_001312236.1, Table 1). Meanwhile, the identification analysis with PCR assay was performed on 302 positive colonies with NIA2 promoter sequences, of which 128 PCR products were sequenced and analyzed, and 3 sequences with biological significance were selected. The 3 sequences were from Populus (XP_006380094.1), Lupinus (CP023132.1), and Nicotiana (XP_016510250.1), respectively (Table 2). Moreover, 3 sequences were selected from 288 positive colonies with 4box sequences, which originate from Gossypium (XM_017753776.1), Ustilaginoidea (KDB15619.1), and Nicotiana (XM_016576899.1, Table 3).
Furthermore, the target plasmids were extracted and validated via electrophoresis. The inserted fragment was 200 bp to 2000 bp in length.
Discussion
Yeast one-hybrid screening is widely recognized as a useful technique for the detection of physical interactions between DNA and DNA-binding proteins, such as TFs [19]. The present study successfully constructed yeast one-hybrid libraries of NR gene (NIA1 and NIA2) promoters and NRE2 cis-element; moreover, several potential TF genes, such as XP_016503563.1, NP_001312236.1, XP_016510250.1, and XM_016576899.1 were screened.
XP_016503563.1 was identified from the NIA1 promoter and annotated in predicted: CRM-domain containing factor CFM3, chloroplastic/mitochondrial-like of Nicotiana, having homology with predicted protein CFM3 in Nicotiana tabacum. The CRM domain is an RNA-binding domain identified in three group II intron splicing factors in chloroplasts, in a family of uncharacterized proteins in plants [20]. Additionally, CRM domain proteins are implicated in the activities of three classes of catalytic RNA, and could interact with catalytic RNA [20]. An in vitro experiment also revealed that the isolated CRM domain in maize has RNA-binding activity [21]. Interestingly, RNA-binding proteins play a key role in the post-transcriptional regulation of gene expression. Besides, some RNA-binding proteins with specific structures can bind to DNA as TFs [22]. CFM3 is a CRM-domain protein related to chloroplast splicing factor CRS1, which dually functions in chloroplast group II intron splicing and mitochondrial gene expression [23]. Therefore, we speculated that XP_016503563.1 may be involved in the transcriptional regulation of the NIA1 promoter in Nicotiana tabacum.
Another gene identified from the NIA1 promoter, NP_001312236.1, was annotated as a sulfite oxidase-like protein in Nicotiana tabacum. Sulfite oxidase is the smallest eukaryotic molybdenum enzyme that utilizes a molybdopterincofactor and aheme group [24]. A study reported that sulfite oxidase and NR have considerable homology, suggesting that they may come from a common family [25]. Notably, Nakamura et al. [26] found that all sulfite oxidases possess a peroxisomal targeting sequence at the C-terminus, and that the peroxisomal targeting sequence could direct polypeptides into peroxisomes in plants, animals, and yeasts. Therefore, the role of sulfite oxidase in transcriptional expression of the NR gene needs to be further verified.
From the NIA2 promoter, 3 TF genes were identified, among which XP_016510250.1 was annotated in PREDICTED: uncharacterized protein LOC107827596 in Nicotiana tabacum. In the Conserved Domain Database, this protein was found to have a pepsin-like aspartic protease domain. Eukaryotic pepsin-like proteases have two domains with similar topological characteristics: C- and N-terminal domains. However, they have limited sequence homology except for the sequences near the active site, indicating that this enzyme may have evolved from ancient copying activity. The active site motif (Asp-Thr/Ser-Gly-Ser) is conserved between retroviruses and eukaryotic proteases, as well as eukaryotic N- and C-terminal pepsin-like proteins. Currently, its relationship with NIA2 promoter gene expression remains to be explored.
XM_016576899.1 was identified from 4 tandem NRE2 cis-elements, which encoded a protein containing RING finger motifs [27]. RING finger proteins contain zinc finger domains consisting of one or several Cys and His residues. Zinc fingers can serve as DNA-binding domains to bind DNA, meeting the structural requirements of TFs. On the basis of the spacing between metal ligands or substitutions at one or more of the metal ligand positions, total 9 RING types were identified: RING-HC, RING-H2, RING-C2, RING-G, RING-mH2, RING-v, RING-D, RING-S/T, and RING-mHC [28]. It was reported that RING finger protein SEVEN IN ABSENTIA OF ARABIDOPSIS2 (SINAT2) may interact with TF RAP2.2 to co-regulate carotenogenesis in Arabidopsis leaves [29]. A recent study revealed that NtRHF1 containing a RING motif in its C-terminal region participated in drought stress response via transcriptional regulation of the downstream stress-responsive genes NtCDPK2, NtLEA5, NtAREB, and NtERD10C in tobacco [30]. Therefore, these results indicate that the RING finger family may regulate the transcriptional expression of genes independently or with other transcription factors.
Conclusions
In conclusion, the identified TF genes from NIA1 and NIA2 promoters and the NRE2 cis-element (such as XP_016503563.1, NP_001312236.1, XP_016510250.1, and XM_016576899.1) may help us to understand the regulatory pathway of nitrate signal response in Nicotiana tabacum. Our results may provide a theoretical basis and possible research directions for investigation of the TFs of the NR gene promoter. Further studies are needed to characterize their functions.
Abbreviations
- ANR1:
-
Arabidopsis nitrate-regulated1
- DSN:
-
Duplex-specific nuclease
- LBD :
-
Lateral organ boundary domain
- LD:
-
Long-distance
- NIR:
-
Nitrite reductase
- NLP7:
-
NIN-like protein 7
- NR:
-
Nitrate reductase
- NRE:
-
Nitrate-responsive cis-element
- OD:
-
Optical density
- PCR:
-
Polymerase chain reaction
- TF:
-
Transcription factor
References
Paungfoo-Lonhienne C, Lonhienne TGA, Rentsch D, Robinson N, Christie M, Webb RI, Gamage HK, Carroll BJ, Schenk PM, Schmidt S. Plants can use protein as a nitrogen source without assistance from other organisms. Proc Natl Acad Sci. 2008;105(11):4524–9.
Chen Q, Mu X, Chen F, Yuan L, Mi G. Dynamic change of mineral nutrient content in different plant organs during the grain filling stage in maize grown under contrasting nitrogen supply. Eur J Agron. 2016;80:137–53.
Chen Y, Xiao C, Wu D, Xia T, Chen Q, Chen F, Yuan L, Mi G. Effects of nitrogen application rate on grain yield and grain nitrogen concentration in two maize hybrids with contrasting nitrogen remobilization efficiency. Eur J Agron. 2015;62(62):79–89.
Vos J, Pelvander P. Effect of nitrogen supply on leaf growth, leaf nitrogen economy and photosynthetic capacity in potato. Field Crop Res. 2005;93(1):64–73.
Scheible WR, Gonzalezfontes A, Lauerer M, Mullerrober B, Caboche M, Stitt M. Nitrate acts as a signal to induce organic acid metabolism and repress starch metabolism in tobacco. Plant Cell. 1997;9(5):783.
Crawford NM, Forde BG. Molecular and developmental biology of inorganic nitrogen nutrition. Arabidopsis Book. 2002;1(1):e0011.
Gojon A, Nacry P, Davidian JC, Salt DE, Williams L. Root uptake regulation: a central process for NPS homeostasis in plants. Curr Opin Plant Biol. 2009;12(3):328–38.
Konishi M, Yanagisawa S. The regulatory region controlling the nitrate-responsive expression of a nitrate reductase gene, NIA1, in Arabidopsis. Plant & Cell Physiology. 2011;52(5):824.
Davenport S, Le LP, Sanchez-Tamburrrino JP. Nitrate metabolism in tobacco leaves overexpressing Arabidopsis nitrite reductase. Plant Physiology & Biochemistry Ppb. 2015;97:96–107.
Suzuki T, Nagai R, Yazaki Y. Mechanisms of transcriptional regulation of gene expression in smooth muscle cells. Circ Res. 1998;82(12):1238–42.
Gan Y, Bernreiter A, Filleur S, Abram B, Forde BG. Overexpressing the ANR1 MADS-box gene in transgenic plants provides new insights into its role in the nitrate regulation of root development. Plant & Cell Physiology. 2012;53(6):1003–16.
Castaings L, Camargo A, Pocholle D, Gaudon V, Texier Y, Boutetmercey S, Taconnat L, Renou JP, Danielvedele F, Fernandez E. The nodule inception-like protein 7 modulates nitrate sensing and metabolism in Arabidopsis. Plant J Cell Mol Biol. 2009;57(3):426–35.
Rubin G, Tohge T, Matsuda F, Saito K, Scheible WR. Members of the LBD family of transcription factors repress anthocyanin synthesis and affect additional nitrogen responses in Arabidopsis. Plant Cell. 2009;21(11):3567.
Konishi M, Yanagisawa S. Identification of a nitrate-responsive cis-element in the Arabidopsis NIR1 promoter defines the presence of multiple cis-regulatory elements for nitrogen response. Plant J Cell Mol Biol. 2010;63(2):269.
Konishi M, Yanagisawa S. Roles of the transcriptional regulation mediated by the nitrate-responsive cis-element in higher plants. Biochem Biophysical Res Commun. 2011;411(4):708.
Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van dPY, Rouzé P, Rombauts S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7.
Jefferson RA. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Report. 1987;5(4):387–405.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72(1–2):248–54.
Luo Y, Vijaychander S, Stile J, Zhu L. Cloning and analysis of DNA-binding proteins by yeast one-hybrid and one-two-hybrid systems. Biotechniques. 1996;20(4):564–8.
Asakura Y, Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant Cell. 2007;19(12):3864.
Barkan A, Klipcan L, Ostersetzer O, Kawamura T, Asakura Y, Watkins K. The CRM domain: an RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13(1):55.
Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. science. 1994;265(5172):615–21.
Asakura Y, Bayraktar OA, Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. Rna. 2008;14(11):2319–32.
Hänsch R, Lang C, Riebeseel E, Lindigkeit R, Gessler A, Rennenberg H, Mendel RR. Plant sulfite oxidase as novel producer of HO. J Biol Chem. 2006;281(10):6884–8.
Kisker C, Schindelin H, Rees DC. Molybdenum-cofactor-containing enzymes: structure and mechanism. Annu Rev Biochem. 1997;66(1):233.
Nakamura T, Meyer C, Sano H. Molecular cloning and characterization of plant genes encoding novel peroxisomal molybdoenzymes of the sulphite oxidase family. J Exp Bot. 2002;53(375):1833–6.
Gmachl M, Gieffers C, Podtelejnikov AV, Mann M, Peters J-M. The RING-H2 finger protein APC11 and the E2 enzyme UBC4 are sufficient to ubiquitinate substrates of the anaphase-promoting complex. Proc Natl Acad Sci. 2000;97(16):8973–8.
Li Y, Wu B, Yu Y, Yang G, Wu C, Zheng C. Genome-wide analysis of the RING finger gene family in apple. Mol Genet Genomics. 2011;286(1):81–94.
Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the Carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007;145(3):1073–85.
Xia Z, Su X, Liu J, Wang M. The RING-H2 finger gene 1 (RHF1) encodes an E3 ubiquitin ligase and participates in drought stress response in Nicotiana tabacum. Genetica. 2013;141(1–3):11–21.
Acknowledgements
None.
Funding
This study was funded by Tobacco Cultivation Key Laboratory of CNTC (grant number YCZP201502). The funding body was not contributed to the design of the study and collection, analysis, and interpretation of data and in writing the manuscript.
Availability of data and materials
All datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Author information
Authors and Affiliations
Contributions
Conception and design of the research: HY, HS; acquisition of data: HY, YZ1; analysis and interpretation of data: YZ; statistical analysis: JW; obtaining funding: HY; drafting the manuscript: HY, HS; revision of manuscript for important intellectual content: JW. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Yang, H., Zhou, Y., Zhang, Y. et al. Identification of transcription factors of nitrate reductase gene promoters and NRE2 cis-element through yeast one-hybrid screening in Nicotiana tabacum. BMC Plant Biol 19, 145 (2019). https://doi.org/10.1186/s12870-019-1724-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-019-1724-z