Abstract
Background
Road-killed wild animals have been classified as sentinels for detecting such zoonotic pathogens as Leishmania spp., offering new opportunities for epidemiological studies of this infection.
Methods
This study aimed to evaluate the presence of Leishmania spp. and Leishmania chagasi DNA by PCR in tissue samples (lung, liver, spleen, kidney, heart, mesenteric lymph node and adrenal gland) from 70 road-killed wild animals.
Results
DNA was detected in tissues of one Cavia aperea (Brazilian guinea pig), five Cerdocyon thous (crab-eating fox), one Dasypus septemcinctus (seven-banded armadillo), two Didelphis albiventris (white-eared opossum), one Hydrochoerus hydrochoeris (capybara), two Myrmecophaga tridactyla (giant anteater), one Procyon cancrivorus (crab-eating raccoon), two Sphiggurus spinosus (porcupine) and one Tamandua tetradactyla (lesser anteater) from different locations in the Central Western part of São Paulo state. The Leishmania chagasi DNA were confirmed in mesenteric lymph node of one Cerdocyon thous. Results indicated common infection in wild animals.
Conclusions
The approach employed herein proved useful for detecting the environmental occurrence of Leishmania spp. and L. chagasi, as well as determining natural wild reservoirs and contributing to understand the host-parasite interaction.
Similar content being viewed by others
Background
Leishmaniosis is a zoonotic, parasitic disease caused by kinetoplastid flagellate protozoan parasites of the genus Leishmania that infects several mammal species, including humans, and is transmitted by the phlebotomine sandfly. Leishmania species include visceral, cutaneous and mucocutaneous forms of the disease in both the Old and New Worlds [1, 2].
Great concern has been sparked by the contribution that global warming might be making to the recent increase in the number of reported cases and geographical areas [3]. Environmental, demographic and human behavioral factors contribute to the changing landscape of leishmaniasis, which includes increased risk factors for zoonotic cutaneous leishmaniasis and new scenarios associated with the zoonotic visceral leishmaniasis [4].
Studies on Leishmania spp. in wild animals have become more numerous in Brazil due to the importance of these species in the life cycle of leishmaniasis [5]. Studies involving road-killed instead of laboratory research animals have become more frequent in helminthological, epidemiological, morphological and genetic areas [6–12]. However, the use of molecular techniques for detection of microorganisms in these samples is recent [13–17].
Despite the difficulty of culturing and histopathologically analyzing tissue samples from road-killed animals, molecular techniques can be used in the identification and typing of pathogens by polymerase chain reaction (PCR), which presents high specificity and sensitivity to a certain fragment of the pathogen’s specific DNA [18].
Leishmania kinetoplastic DNA (kDNA)-specific probes have been used for the detection and identification of this protozoan, and demonstrated to be useful for epidemiological field studies because a large number of samples can be handled simultaneously [19]. A minicircle of kDNA (0.8 to 1 kb in length) is an ideal target, since it is present in 10,000 copies per cell and because its sequences are known for most Leishmania species [20].
The present work aimed to describe possible new hosts for leishmaniasis by using molecular tools to detect Leishmania spp. in tissues of road-killed wild mammals. Thus, research into new hosts by molecular techniques is distinctive in epidemiological studies of pathogens and represents suitable indicators of environmental contamination by Leishmania spp. [21].
Methods
Animals and studied area
Seventy road-killed wild animals were studied: one Callithrix penicillata (black-tufted marmoset), four Cavia aperea (Brazilian guinea pig), one Cebus apella (capuchin monkey), 13 Cerdocyon thous (crab-eating fox), three Dasypus novemcinctus (nine-banded armadillo), one Dasypus septemcinctus (seven-banded armadillo), nine Didelphis albiventris (white-eared opossum), one Eira barbara (tayra), one Euphractus sexcinctus (six-banded armadillo), two Gallictis vittata (grison), two Hydrochoerus hydrochaeris (capybara), one Leopardus tigrinus (leopard cat), five Lepus europaeus (brown hare), three Lutreolina crassicaudata (latrine opossum), two Mazama gouazoubira (brown brocket deer), one Myocastor coypus (coypu), six Myrmecophaga tridactyla (giant anteater), three Procyon cancrivorus (crab-eating raccoon), two Puma concolor (cougar), two Rattus rattus (black rat), five Sphiggurus spinosus (porcupine) and two Tamandua tetradactyla (lesser anteater). Only recently killed animals (1–7 hours) and those with no exposed viscera were collected. This study is in accordance with the Brazilian Institute of Environment and Renewable Natural Resources’ (IBAMA) normative statement n. 119 of October 11, 2006, chapter VI, art .26, which authorizes the collection and transport of animals that were found dead for scientific or didactic purposes. This work was also approved by the Ethics Committee for Animal Experimentation at our Institution (CEEA/FMVZ n.211/2008).
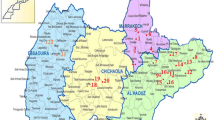
The geographic positions of the road-killed animals, established through global positioning system (GPS), were plotted on a digital map using a geographic database by the TerraView 3.6.0 [22].
Molecular detection
DNA extraction from the animals’ tissue samples (lung, spleen, liver, kidney, heart, mesenteric lymph node and adrenal gland) was carried out by using the kit Illustra™ Tissue & Cells Genomic Prep Mini Spin (GE Healthcare, USA). PCR reactions were performed by employing the primers LinR4 (5’-GGGTTGGTGTAAAATAGGG-3’) and Lin19 (5’-CAGAACGCCCCTACCCG-3’), described by Aransay et al. [20], to amplify a 720 bp fragment. Samples positive for Leishmania spp. PCR were also assayed for Leishmania braziliensis complex and Leishmania mexicana complex [23, 24]. Genus-specific primers for Leishmania spp. were used in order to identify the DNA of all possible Leishmania species that cause visceral or cutaneous leishmaniasis. The cycling profile consisted of an initial denaturation at 95°C for three minutes, followed by 30 cycles at 95°C for 30 seconds, 63°C for 30 seconds and 72°C for one minute, and a final extension at 72°C for seven minutes. Positive controls were included in each assay and consisted of 10 ng of DNA extracted from Leishmania major (MHOM/SU/1973/5-ASKH) and L. chagasi (MHOM/BR/2002/LPC-RPV). Negative controls were: ultrapure water and DNA from T. cruzi (ColTryp 0032/MCAN/BR/2008/CAO) that were added to the mix-PCR. The PCR mixture was composed of 10 mM Tris HCl pH 8.0, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP, 10 ρmol of each primer, 0.2 units of Taq DNA polymerase, and 10 ng DNA template.
The amplification of Leishmania chagasi DNA was performed utilizing primers Lc14 (5’-CGCACGTTATATCTACAGGTTGAG-3’) and Lc15 (5’- TGTTTGGGATTGAGGTAATAGTGA-3’) on a 190 bp fragment, by using the following cycling profile: initial denaturation at 94°C for four minutes, 40 cycles of 94°C for 30 seconds, 59°C for 30 seconds, 72°C for 30 seconds, and 70°C for ten minutes. Positive controls were included in each assay and consisted of 10 ng of DNA extracted from L. chagasi (MHOM/BR/2002/LPC-RPV). Negative controls were: ultrapure water and DNA from T. cruzi (ColTryp 0032/MCAN/BR/2008/CAO) that were added to the mix-PCR. The PCR mixture was composed of 10 mM Tris HCl pH 8.0, 50 mM KCl, 1.5 mM MgCl2, 0.2 mM dNTP, 10 ρmol of each primer, 0.5 units of Taq DNA polymerase, and 10 ng DNA template.
Amplification was performed in a MasterCycler EP gradient (Eppendorf, USA). The sequence was analyzed by electrophoresis in 1.5% agarose with SYBR® safe DNA gel stained (Invitrogen, USA), and visualized in an image analyzer (GelDoc-It™ Imaging System – UVP, USA) by using VisonWorks®LS Software. Amplicons were purified by using ExoSap (USB, USA) and the sequencing reactions were carried out on both strands in a 3500 Genetic Analyzer (Applied Biosystems). The obtained sense and antisense sequences were visualized (Chromas 2.3 software, Technelysium Pty Ltd, Australia), aligned by the software MEGA 4 and compared with the NCBI database using BLASTn (Basic Local Alignment Tool for Nucleotide) [25].
Results and discussion
The present results draw attention to a very important source of research and emphasize the importance of using this biological resource in an epidemiological study of zoonotic infection.
One of the main problems in elucidating leishmaniasis epidemiology is to identify and confirm that a vertebrate host is a natural reservoir. The natural reservoirs are widely unknown because of the difficulties in capturing a sufficient number of wild animals and due to the techniques used in isolating and identifying the parasite.
This approach of using road-killed wild animals for the molecular detection of Leishmania spp. may represent a useful alternative to the utilization of captured ones in research studies, as indicated by animal research ethics committees. In the present study, a great diversity of road-killed wild mammal species was found. Culture analysis and histopathology are difficult and laborious. Sensitive and specific molecular tools allow pathogens to be identified without the need of culturing.
In this paper, molecular detection of Leishmania spp. and Leishmania chagasi was attempted from several wildlife species using PCR. Several studies have reported the presence of this parasite in mammalian species, including rodents, carnivores, primates and marsupials [5, 26–29].
Table 1 contains the results of the PCR and identifies percentages of amplicon obtained in road-killed wild animals positive for Leishmania spp. and Leishmania chagasi from deposited homologue DNA sequences, as determined by BLASTn analysis. Figure 1 illustrates the human cutaneous leishmaniasis data corresponding to the cases seen in the central western area of the state of São Paulo, Brazil, from 1998 to 2010 [30]. Figure 2 displays the human visceral leishmaniasis data corresponding to the cases seen in the central western area of the state of São Paulo, Brazil, from 1998 to 2010 and geographic location of the positive road-killed animals evaluated [31].
Leishmania DNA was detected in 5/12 (41.67%; CI95% 19.22-68.42%) samples from Cerdocyon thous (crab-eating fox). A previous report indicates seropositivity in wild non-captive Cerdocyon thous[5, 28]. The importance of these animals as reservoirs depends on their ability to transmit the infection to sandflies rather than on their infection rate; it is also a function of their capability to (re)introduce the pathogen into Leishmania-free dog populations [32].
In the current study, Leishmania spp. DNA was detected in 1/3 Procyon cancrivorus (crab-eating raccoon). Voltarelli et al.[33] reported the presence of Leishmania antibodies in Procyon cancrivorus in Northwestern Paraná. These findings suggest that these species can act as a reservoir for Leishmania spp.
The members of Didelphidae, represented by Didelphis albiventris specimens (white-eared opossums), are habitat generalists and currently occur in areas near dwellings, including farms, yards and urban centers [34]. This species is already proven to be a leishmaniasis reservoir and, for its synanthropic habits, it plays an important role in the peridomestic-forest traffic of degraded areas [35–37]. The present study confirms that molecular detection of Leishmania spp. in 2/8 group members may be common, as was already described in several regions of Brazil: Manaus, Amazonas state; in Barra de Guarituba, Rio de Janeiro state; in Amaraji, Pernambuco; and Bauru, São Paulo state [38–41].
Rodents were represented by five S. spinosus (porcupine), four C. aperea (Brazilian guinea pig), two Rattus rattus (black rat), two Hydrochoerus hydrochaeris (capybara) and one M. coypus (coypu). Current data show that 8/14 (57.14%; CI95% 35.14-82.34%) specimens of wild rodents were positive, a finding that corroborates the literature that considered some rodent as reservoirs of Leishmania spp.
The superorder Xenarthra was represented by 13 specimens: three D. novemcinctus (nine-banded armadillo), one E. sexcinctus (six-banded armadillo), one D. septemcinctus (seven-banded armadillo), two T. tetradactyla (lesser anteater) and six M. tridactyla (giant anteater). These animals present some peculiar physiological and ecological characteristics including a weak immune system and low body temperature, besides the fact that they live literally immersed in soil and organic matter, mainly in tropical and subtropical regions, under biotic and abiotic conditions that promote multiple encounters with a diverse group of pathogens and vectors.
The present study confirms the occurrence of Leishmania spp. DNA in armadillos (one D. septemcintus) and anteaters (one T. tetradactyla and two M. tridactyla). Casadeval and Pirofski [42] clarified many points on virulence and pathogenicity regarding host immune response and pathogen activity. According to the authors, there are classes of pathogenic microorganisms varying from those that provoke damage in hosts that present an extremely weak immune response to others that cause disease only in a situation of very strong immune response. Therefore, it seems reasonable to consider Leishmania spp. to be a pathogen whose ability to provoke disease also depends on host immune response. Since the cellular immune response is weak in armadillos and anteaters, it is possible to detect yeast cells in many of their organs; however, this is not sufficient to cause disease as observed in human hosts. Taken together, these factors make xenarthrans suitable models for studying host-pathogen interaction [43].
These animals are assumed to be sources of infection since the agent’s DNA was found in internal organs; in addition, parasitism may occur in internal and cutaneous organs, facilitating transmission from the blood meal by the vector that inoculates promastigote forms of the agent into the man while sucking.
The identities of the amplicon were confirmed by direct double-strand sequencing which showed 100% similarity with L. chagasi sequence deposited at GenBank (access number AF308682.1) (Table 1).
Even without the DNA detection of the cutaneous leishmaniasis agents, the positive results for Leishmania spp. are interesting. Of the 20 species described in the New World, five have never been reported to have caused visceral human leishmaniasis: Leishmania enriettii, Leishmania hertigi, Leishmania deanei, Leishmania aristidesi and Leishmania forattinii[44–48]. The L. forattinii was isolated from pooled liver and spleen of opossum Didelphis marsupialis captured in Conchas, SP, Brazil [48, 49].
It is suggested that the species is Leishmania forattinii and that the evaluated site is close to that where the parasite was first isolated, since the species nucleotide sequence deposited at the GenBank was not found. Considering the occurrence of both the cutaneous and visceral form, in the studied municipalities, it must be emphasized that the sandfly vector may be present and serve as transmitter of Leishmania to these animals and humans.
These findings corroborate the worldwide distribution of Leishmania spp., considering the wide variety of intermediate hosts that contribute to the epidemiological transmission chain of this infection.
It is important to emphasize that Bauru, SP, is endemic for leishmaniasis; therefore, our results indicate the need for epidemiological molecular biology research on environmental contamination by L. chagasi.
It was possible to evaluate 22 different wild species, without the necessity of exerting a laborious sampling effort. In fact, the numbers and diversity of road-killed animals are considerably higher and, in general, they are killed after their own natural habitats had been invaded by roads [50]. In this manner, the geographic coordinates of the locations of the infected animals are well-integrated in databases that use the geographical information systems (GIS), thus contributing to a better understanding of pathogen distribution.
These results show risk factors such as free movement of the circulating parasite and vectors, as well as the importance of road-killed animals as possible reservoirs for the transmission of Leishmania spp. in addition to the significance of the environment and ecology of these positive mammals in the interaction of Leishmania with different Leishmania species that may be pathogenic to humans.
Conclusions
The presented results focus that road-killed animals may serve as an important reservoir for transmission of Leishmania spp. and L. chagasi, as well as contributing to understand the host-parasite interaction.
References
Gontijo B, Carvalho MLR: Leishmaniose tegumentar americana. Rev Soc Bras Med Trop 2003, 36(1):71–80.
Gontijo CMF, Melo MN: Leishmaniose visceral no Brazil: quadro atual, desafios e perspectivas. Rev Bras Epidemiol 2004, 7(3):338–349. 10.1590/S1415-790X2004000300011
Desjeux P: Leishmaniasis: currents situation and new perspectives. Comp Immunol Microbiol Infect Dis 2004, 27(5):305–318. 10.1016/j.cimid.2004.03.004
Gramiccia M, Gradoni L: The current status of zoonotic leishmaniases and approaches to disease control. Int J Parasitol 2005, 35(11–12):1169–1180.
Luppi MM, Malta MC, Silva TM, Silva FL, Motta RO, Miranda I, Ecco R, Santos RL: Visceral leishmaniasis in captive wild canids in Brazil. Vet Parasitol 2008, 155(1–2):146–151.
Coyner DF, Wooding JB, Forrester DJ: A comparison of parasitic helminths and arthropods from two subspecies of fox squirrels ( Sciurus niger ) in Florida. J Wildl Dis 1996, 32(3):492–497. 10.7589/0090-3558-32.3.492
Cheadle MA, Tanhauser SM, Dame JB, Sellon DC, Hines M, Ginn PE, Mackay RJ, Greiner EC: The nine-banded armadillo ( Dasypus novemcinctus ) is an intermediate host for Sarcocystis neurona . Int J Parasitol 2001, 31(4):330–335. 10.1016/S0020-7519(01)00177-1
Foster GW, Main MB, Kinsella JM, Dixon LM, Terrell SP, Forrester DJ: Parasitic helminths and arthropods of coyotes ( Canis latrans ) from Florida. USA Comp Parasitol 2003, 70(2):162–166. 10.1654/4081
Nelder MP, Reeves WK: Ectoparasites of road-killed vertebrates in northwestern South Carolina, USA. Vet Parasitol 2005, 129(3–4):313–322.
Ferroglio E, Ragagli C, Trisciuoglio A: Physaloptera sibirica in foxes and badgers from the Western Alps (Italy). Vet Parasitol 2009, 163(1–2):164–166.
Hoppe EG, Araújo de Lima RC, Tebaldi JH, Athayde AC, Nascimento AA: Helminthological records of six-banded armadillos Euphractus sexcinctus (Linnaeus, 1758) from the Brazilian semi-arid region, Patos county, Paraíba state, including new morphological data on Trichohelix tuberculata (Parona and Stossich, 1901) Ortlepp, 1922 and proposal of Hadrostrongylus ransomi nov. comb. Braz J Biol 2009, 69(2):423–428. 10.1590/S1519-69842009000200027
Miquel J, Foronda P, Torres J, Swiderski Z, Feliu C: Ultrastructural study of the spermatozoon of Taenia taeniaeformis (Batsch, 1786) (Cestoda, Cyclophyllidea, Taeniidae), an intestinal parasite of Felis catus from La Palma (Canary Islands, Spain). Parasitol Res 2009, 104(6):1477–1483. 10.1007/s00436-009-1351-y
Richini-Pereira VB, Bosco SMG, Griese J, Theodoro RC, Macoris SA, da Silva RJ, Barrozo L, Tavares PM, Zancopé-Oliveira RM, Bagagli E: Molecular detection of Paracoccidioides brasiliensis in road-killed wild animals. Med Mycol 2008, 46(1):35–40. 10.1080/13693780701553002
Richini-Pereira VB, Bosco SMG, Theodoro RC, Barrozo L, Pedrini SCB, Rosa PS, Bagagli E: Importance of xenarthrans in the eco-epidemiology of Paracoccidioides brasiliensis . BMC Res Notes 2009, 2: 1–6. 10.1186/1756-0500-2-1
Richini-Pereira VB, Bosco SMG, Theodoro RC, Barrozo L, Bagagli E: Road-killed wild animals: a preservation problem useful for eco-epidemiological studies of pathogens. J Venom Anim Toxins incl Trop Dis 2010, 16(4):607613. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1678–91992010000400011
Zhao C, Onuma M, Asakawa M, Nagamine T, Kuwana T: Preliminary studies on developing a nested PCR assay for molecular diagnosis and identification of nematode ( Heterakis isolonche ) and trematode ( Glaphyrostomum sp.) in Okinawa rail ( Gallirallus okinawae ). Vet Parasitol 2009, 163(1–2):156–160.
Pedrini SC, Rosa PS, Medri IM, Mourão G, Bagagli E, Lopes CA: Search for Mycobacterium leprae in wild mammals. Braz J Infect Dis 2010, 14(1):47–53. 10.1016/S1413-8670(10)70010-6
Persing DH, Smith TF, Tenover FC, White TJ: Diagnostic Molecular Microbiology: Principles and Applications. Washington, D.C: American Society for Microbiology; 1993:423–430.
Esseghir SA, Ftaiti A, Ready PB, Khdraoui B, Zaafouri K, Dellagi K, Ben Ismail R: The squash blot technique and the detection of Leishmania major in Phlebotomus papatasi in Tunisia. Arch Inst Pasteur Tunis 1993, 70(3–4):493–496.
Aransay AM, Scoulica E, Tselentis Y: Detection and identification of Leishmania DNA within naturally infected sand flies by seminested PCR on minicircle kinetoplastic DNA. Appl Environ Microbiol 2000, 66(55):1933–1938.
Gennari SM, Canón-Franco WA, Yai LE, de Souza SL, Santos LC, Farias NA, Ruas J, Rossi FW, Gomes AB: Seroprevalence of Toxoplasma gondii antibodies from wild canids from Brazil. Vet Parasitol 2004, 121(3–4):337–340.
INPE: Instituto Nacional de Pesquisas Espaciais. TerraView: Software; v.3.6.0 [http://www.dpi.inpe.br/terraview/index.php]
de Bruijn MH, Barker DC: Diagnosis of New World leishmaniasis: specific detection of species of the Leishmania braziliensis complex by amplification of kinetoplast DNA. Acta Trop 1992, 52(1):45–58. 10.1016/0001-706X(92)90006-J
Eresh S, McCallum SM, Barker DC: Identification and diagnosis of Leishmania mexicana complex isolates by polymerase chain reaction. Parasitololy 1994, 109(Pt 4):423–433.
Tamura K, Dudley J, Nei M, Kumar S: MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007, 24(8):1596–1599. 10.1093/molbev/msm092
Alexander B, Lozano C, Barker DC, McCann SH, Adler G: Detection of Leishmania ( Viannia ) braziliensis complex in wild mammals from Colombian coffee plantations by PCR and DNA hybridization. Acta Trop 1998, 69(1):41–50. 10.1016/S0001-706X(97)00114-9
Travi BL, Osorio Y, Becerra MT, Adler GH: Dynamics of Leishmania chagasi infection in small mammals of the undisturbed and degraded tropical dry forests of northern Colombia. Trans R Soc Trop Med Hyg 1998, 92(3):275–278. 10.1016/S0035-9203(98)91009-4
Curi NHA, Miranda I, Talamoni AS: Serologic evidence of Leishmania infection in free-ranging wild and domestic canids around a Brazilian National Park. Mem Inst Oswaldo Cruz 2006, 101(1):99–101. 10.1590/S0074-02762006000100019
Papadogiannakis E, Spanakos G, Kontos V, Menounos PG, Tegos N, Vakalis N: Molecular detection of Leishmania infantum in wild rodents ( Rattus norvegicus ) in Greece. Zoonoses Public Health 2010, 57(7–8):23–25.
Centro de Vigilância Epidemiológica: Leishmaniose tegumentar americana. Distribuição do número de casos de Leishmaniose Tegumentar por município provável de infecção. Estado de São Paulo, 1998–2012. http://www.cve.saude.sp.gov.br/htm/zoo/lta_lpi.htm
Centro de Vigilância Epidemiológica: Leishmaniose visceral americana. Distribuição do número de casos e óbitos de LVA segundo município e GVE de infecção. Estado de São Paulo 1998–2011. http://www.cve.saude.sp.gov.br/htm/zoo/lvah_lpi.htm
Dressen DW: Toxoplasma gondii infections in wildlife. J Am Vet Med Assoc 1990, 196: 274–276.
Voltarelli EM, Arraes SMAA, Perles TF, Lonardoni MVC, Teodoro U, Silveira TGV: Serological survey for Leishmania sp. infection in wild animals from the municipality of Maringá, Paraná State, Brazil. J Venom Anim Toxins incl Trop Dis 2009, 15(4):732–744. http://www.scielo.br/scielo.php?script=sci_arttext&pid=S1678–91992009000400011 10.1590/S1678-91992009000400011
Câmara T, Murta R: Mamíferos da Serra do Cipó. PUC, Minas, Museu de Ciências Naturais: Belo Horizonte; 2003.
Forrester DJ: Parasites and Diseases of Wild Mammals in Florida. First edition. Gainesville: University Press of Florida; 1992:459.
Guerra JAO, Ribeiro JAS, Coelho LIARC, Barbosa MGV, Paes MG: Epidemiologia da leishmaniose tegumentar na comunidade São João, Manaus, Amazonas. Brasil Cad Saúde Pública 2006, 22(11):2319–2327.
Quintal AP, Ribeiro Ede S, Rodrigues FP, Rocha FS, Floeter-Winter LM, Nunes CM: Leishmania spp. in Didelphis albiventris and Micoureus paraguayanus (Didelphimorphia: Didelphidae) of Brazil. Vet Parasitol 2011, 176(2–3):112–119.
Arias JR, Naiff RD: The principal reservoir host of cutaneous leishmaniasis in the urban areas of Manaus, Central Amazon of Brazil. Mem Inst Oswaldo Cruz 1981, 76(3):279–286.
Cabrera MAA, Paula AA, Camacho LAB, Marzochi MCA, Xavier SC, da Silva AVM, Jansen AM: Canine visceral leishmaniasis in Barra de Guaratiba, Rio de Janeiro, Brazil: assessment of risk factors. Rev Inst Med Trop Sao Paulo 2003, 45(2):79–83. 10.1590/S0036-46652003000200005
Brandão-Filho SP, Brito ME, Carvalho FG, Ishikawa EA, Cupolillo E, Floeter-Winter L, Shaw JJ: Wild and synanthropic host of Leishmania (Viannia) braziliensis in the endemic cutaneous leismaniasis locality of Amaraji, Pernambuco State, Brazil. Trans R Soc Trop Med Hyg 2003, 97(3):291–296. 10.1016/S0035-9203(03)90146-5
Santiago ME, Vasconcelos RO, Fattori KR, Munari DP, Michelin Ade F, Lima VM: An investigation of Leishmania spp. in Didelphis spp. from urban and peri-urban areas in Bauru (São Paulo, Brazil). Vet Parasitol 2007, 150(4):283–290. 10.1016/j.vetpar.2007.09.026
Casadevall A, Pirofski LA: Host-pathogen interactions: redefining the basic concepts of virulence and pathogenicity. Infect Immun 1999, 67(8):3703–3713.
Bagagli E, Bosco SMG: Armadillos and dimorphic pathogenic fungi: ecological and evolutionary aspects. In The Biology of the Xenarthra. First edition. Edited by: Viscaino SF, Loughry WJ. Gainesville: University Press of Florida; 2008:103–110.
Muniz J, Medina HSG: Leishmaniose tegumentar do cobaio (Leishmania enriettii) . Arq Biol Tecnol 1948, 3(2):7–25.
Herrer A: Leishmania hertigi sp. n., from the tropical porcupine, Coendou rothschildi Thomas . J Parasitol 1971, 57(3):626–629. 10.2307/3277928
Lainson R, Shaw JJ: Leishmanias of neotropical porcupines: Leishmania hertigi deanei nov. subsp. Acta Amaz 1977, 7(1):51–57.
Lainson R, Shaw JJ: The role of animals in the epidemiology of South American leishmaniasis. In The Biology of the Kinetoplastida, Volume 2. Edited by: Lumsden WHR, Evans DA. London, New York, San Francisco: Academic Press; 1979:1–116.
Yoshida ELA, Silva R, Cortez LS, Corrêa FMA: Encontro de espécie do gênero Leishmania em Didelphis marsupialis aurita no Estado de São Paulo, Brasil. Rev Inst Med Trop Sao Paulo 1979, 21: 110–113.
Yoshida ELA, Cuba CA, Pacheco RS, Cupolillo E, Tavares CC, Machado GMC, Momen H, Grimaldi JG: Description of Leishmania (Leishmania) forattinii sp. n., a new parasite infecting opossums and rodents in Brazil. Mem Inst Oswaldo Cruz 1993, 88(3):397–406. 10.1590/S0074-02761993000300008
Laurance WF, Goosem M, Laurance SG: Impacts of roads and linear clearings on tropical forests. Trends Ecol Evol 2009, 24(12):659–669. 10.1016/j.tree.2009.06.009
Acknowledgments
The authors would like to thank the State of São Paulo Research Foundation (FAPESP, grants n. 08/09378-8 and n. 08/08291-6) for their funding of this research.
Ethics committee approval
This study is in accordance with the Brazilian Institute of Environment and Renewable Natural Resources’ (IBAMA) normative statement n. 119 of October 11, 2006, chapter VI, art .26, which authorizes the collection and transport of animals that were found dead for scientific or didactic purposes. This work was also approved by the Ethics Committee for Animal Experimentation of our Institution (CEEA/FMVZ n.211/2008).
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interests
The authors declare that there are no competing interests.
Authors’ contributions
VBRP participated in the design of the study, data collection, laboratory tests, analysis and interpretation of data, writing and editing of the manuscript. PMM and EYH participated in the data collection, laboratory tests and took part in the writing. CV participated in geographical location of animals, analysis and interpretation of data, writing and revision of the manuscript. RCS participated in the analysis and interpretation of data, writing and revision. HL was responsible for the coordination, study design, analysis and interpretation of data, writing and editing of the manuscript. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Richini-Pereira, V.B., Marson, P.M., Hayasaka, E.Y. et al. Molecular detection of Leishmania spp. in road-killed wild mammals in the Central Western area of the State of São Paulo, Brazil. J Venom Anim Toxins Incl Trop Dis 20, 27 (2014). https://doi.org/10.1186/1678-9199-20-27
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1678-9199-20-27