Abstract
Background
Abnormal blood glucose (BG) concentrations have been associated with increased morbidity and mortality in both critically ill adults and infants. Furthermore, hypoglycaemia and glycaemic variability have both been independently linked to mortality in these patients. Continuous Glucose Monitoring (CGM) devices have the potential to improve detection and diagnosis of these glycaemic abnormalities. However, sensor noise is a trade-off of the high measurement rate and must be managed effectively if CGMs are going to be used to monitor, diagnose and potentially help treat glycaemic abnormalities.
Aim
To develop a tool that will aid clinicians in identifying unusual CGM behaviour and highlight CGM data that potentially need to be interpreted with care.
Methods
CGM data and BG measurements from 50 infants at risk of hypoglycaemia were used. Unusual CGM measurements were classified using a stochastic model based on the kernel density method and historical CGM measurements from the cohort. CGM traces were colour coded with very unusual measurements coloured red, highlighting areas to be interpreted with care. A 5-fold validation of the model was Monte Carlo simulated 25 times to ensure an adequate model fit.
Results
The stochastic model was generated using ~67,000 CGM measurements, spread across the glycaemic range ~2-10 mmol/L. A 5-fold validation showed a good model fit: the model 80% confidence interval (CI) captured 83% of clinical CGM data, the model 90% CI captured 91% of clinical CGM data, and the model 99% CI captured 99% of clinical CGM data. Three patient examples show the stochastic classification method in use with 1) A stable, low variability patient which shows no unusual CGM measurements, 2) A patient with a very sudden, short hypoglycaemic event (classified as unusual), and, 3) A patient with very high, potentially un-physiological, glycaemic variability after day 3 of monitoring (classified as very unusual).
Conclusions
This study has produced a stochastic model and classification method capable of highlighting unusual CGM behaviour. This method has the potential to classify important glycaemic events (e.g. hypoglycaemia) as true clinical events or sensor noise, and to help identify possible sensor degradation. Colour coded CGM traces convey the information quickly and efficiently, while remaining computationally light enough to be used retrospectively or in real-time.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
Abnormal blood glucose (BG) concentrations have been associated with increased morbidity and mortality in both critically ill adults and infants. Patients in the intensive care unit (ICU) often experience high levels of insulin resistance [1–7] and stress-induced hyperglycaemia, which can negatively impact outcomes [1–3, 7, 8]. Similarly, hyperglycaemia in very-low-birth-weight or premature infants is also associated with negative outcomes [9, 10]. Further complicating the matter, hypoglycaemia and glycaemic variability have both been independently linked to mortality in critically ill patients [11–14].
Diagnosis of hyperglycaemia and hypoglycaemia in critically ill patients is by BG measurements, which are typically taken several hours apart. More frequent BG measurements are not clinically practical due to the additional nursing workload [15–17] and in preterm infants, blood volume considerations. Consequently, important glycaemic events between BG measurements can go undetected. Continuous Glucose Monitoring (CGM) devices have the potential to improve the detection and diagnosis of these glycaemic abnormalities. The continuous glucose monitoring system (CGMS® System Gold™ Medtronic, Minimed, Northridge, CA, USA) provides a glucose value every 5 minutes or 288 measurements per day, with only 2–4 BG measurements per day required for device calibration.
There have been relatively few successful investigations of CGMs in critical care use [18–21], although they are well studied in Type 1 diabetes [22–24]. In particular, one set of tight glycaemic control trials using CGM technology was not particularly successful due, in part, to significant sensor noise [25, 26]. In some cases, added sensor noise is a trade-off for the CGM’s far higher, automated sampling rate [18, 27] and must be effectively managed for these devices to be used successfully. However, these sensor and algorithm technologies are also constantly evolving with every new generation offering improvements [24, 28].
If CGM devices are to be used in the clinical setting to monitor, diagnose and potentially aid in the treatment of abnormal glycaemia, clinicians need to know the data are reliable and accurate. Consider a scenario in which CGM data are retrospectively analysed to classify hypoglycaemia in neonates, where frequent BG measurements are not available. Three consecutive measurements in a CGM trace read 4 mmol/L, 2.5 mmol/L, followed by 4 mmol/L. If hypoglycaemia was classified as a measurement below 2.6 mmol/L, then this would be recorded as a hypoglycaemic event. However, if the rest of the CGM trace was very stable with low variability, intuition would suggest this 'event' is potentially a sensor artefact. There currently is no reliable method to determine which of these interpretations is more likely to be “true”.
Our manuscript describes a tool that will aid clinicians in identifying unusual CGM behaviour, retrospectively or in real-time, and highlight sections of the CGM glucose trace that potentially need to be interpreted with care. Specifically, the focus is to identify unusual CGM sensor behaviour, not unusual glycaemic excursions.
Subjects and methods
Subjects
This study used CGM data from 50 babies at risk of hypoglycaemia who were admitted to the Waikato Hospital Newborn Intensive Care Unit (NICU). The cohort contained 26 males and 24 females with a median gestational age of 34 weeks and a median birth weight of 2172 g. The primary risk factors used to identify infants likely to become hypoglycaemic include diabetic mother, prematurity and being small or large for gestational age. The study was approved by the Northern Y Regional Ethics Committee.
Continuous glucose monitoring
All patients had interstitial glucose monitoring using the CGMS® System Gold™ (Medtronic, Minimed, Northridge, CA, USA). Monitoring began on admission to the NICU and finished after 7 days or when the baby was no longer considered to be at risk of hypoglycaemia, whichever came first. During the monitoring period nurses were asked to record all BG concentrations, feeding and medication for the management of hypoglycaemia. However, they remained blind to the glucose concentrations determined by the device. The device was calibrated per the manufacturer’s recommendations and all of the data entered into the device were checked against clinical records for accuracy. Upon completion of monitoring, data were downloaded to a PC using CGMS system solutions software version 3.0C, which calibrated the CGM readings retrospectively. A total of 234 days of CGM data (67438 measurements) were collected and the per-patient median [IQR (inter-quartile range)] duration of monitoring was 4.7 [4.0 - 5.7] days.
Calibration measurements
The CGM device required calibration 2–4 times daily to convert the electrical current produced by the sensor into a meaningful glucose value. Blood samples were taken by nursing staff via heel-pricking and the glucose concentration was used to calibrate the CGM device. The median [inter-quartile range (IQR)] interval between samples was 4.8 [3.5 – 6.4] hours.
All BG calibration measurements were made using a blood gas analyser (Radiometer, ABL800Flex, Copenhagen) using the glucose oxidase method. This device has a reading range of 0.0 to 60.0 mmol/L and a C.V. of 1.4-2.2% [29, 30]. Furthermore, a study by Watkinson et al. showed that a device from the same family, using the same glucose electrode, had a coefficient of variation of 2.1% in ICU patients and performance was not affected by haematocrit, pH or PaO2 [30]. Due to the location of the blood gas analyser, a short time delay (estimated < 15mins maximum) was possible between taking the blood sample and introducing the resulting measurement into the device.
Stochastic model
A stochastic model based on the kernel density method was implemented to classify unusual CGM measurements using the previous CGM measurement and information about the history of CGM behaviour. The model is an extension to the methods described by Lin et al. [27] who developed a stochastic model for insulin sensitivity prediction.
Where: and
The model was generated using Equations 1 and 2 which define the 2-dimensional kernel density estimation in conditional CGM measurement variability. Each and is a normal probability density function (pdf) centred at the corresponding xi and yi. To force non-negativity in x and y, and normalise each and in the positive domain. Specifically, for a given CGM measurement CGM n-1 , Equations 1 provides a continuous, empirical estimate of the conditional pdf for the next CGM measurement, CGM n . These conditional pdf’s provide the basis for classifying CGM measurements and identifying unusual CGM behaviour.
CGM measurement classification
Using the stochastic model, a given CGM measurement, CGM n , is classified as follows:
-
1.
The previous measurement, CGM n-1 , is used to find the corresponding conditional pdf from the model.
-
2.
CGM n is located in the pdf and its percentile value in the conditional pdf is determined.
-
3.
The percentile is used to classify CGM n , where a very high or very low percentile is indicative of an outlier. These outliers are classified as unusual CGM measurements.
The measurement-to-measurement sections of the CGM trace were colour coded based on the percentile value, to highlight areas of unusual CGM behaviour quickly and effectively. Three confidence intervals (CI’s) were used to specify the colour: within 80% CI (10th-90th percentile) was blue, within 90% CI (5th-95th percentile) was cyan, within 99% CI (0.5th-99.5th percentile) was yellow, and outside 99% CI was red. These intervals were chosen based on the data used in this study and can be customised for different patient groups and/or different CGM sensors. As the scale starts at 80% CI, the focus here is on classifying outliers, rather than the full range.
5-fold validation of stochastic model
A 5-fold validation was used to check the fit of the stochastic model. The data set (N = 50) was randomly divided into 5 sets of 10 patients. For each 10 patient group, the remaining 40 patients were used to create a stochastic model which was then tested on the group of 10 patients. The model fit was assessed by counting the number of clinical CGM measurements (from the 10 patients) captured by the model’s 80% CI, 90% CI and 99% CI.
Due to the random nature of this validation procedure, Monte Carlo (MC) methods were used to reduce the effect of randomly selected outliers on overall results. MC methods provide a robust means of estimating the range of possible outcomes for a process involving one or more random variables. In this study, the MC simulation involved repeating the 5-fold validation 25 times, and reporting the Median [IQR] results.
Results
Clinical CGM data and stochastic model generation
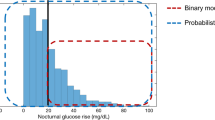
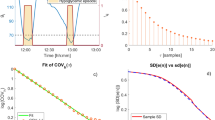
Figure 1 shows a plot of all the CGM data (CGM n-1 , CGM n ). The contour lines represent the 5th, 25th, 50th, 75th, and 95th percentiles of the stochastic model surface. Figure 2 shows a distribution of the data density by glycaemic level. Figure 3 shows a surface plot of the stochastic model. Conditional probability density functions are slices parallel to the CGM n axis, and each slice has an area under the curve of 1.0. Figure 4 shows a comparison of the pdf's obtained from the model versus the pdf's obtained directly from the CGM data. Each pdf shows the expected distribution of CGM n given a previous measurement (CGM n-1 ) of 2, 4, 6, 8 or 10 mmol/L. It should be noted that the pdf's could be generated for any value of CGM n-1 within the bounds of the model; Figure 4 shows just five examples.
Stochastic model surface. Data density by glycaemic level. Stochastic model surface for this data set. Conditional probability density functions are the surface slices along CGMn-1axis, each slice has an area under the curve summing to 1.0. A colour gradient was used to show the height of the surface.
Classification of representative CGM data
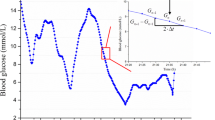
Figures 5, 6, 7 show three examples of CGM traces that have been coloured using the stochastic classification method. Figure 5 shows a stable trace, which is almost entirely dark blue, indicating the measurement-to-measurement change throughout the trace is not unusual. Figure 6 shows a trace with several potentially unusual measurements throughout the trace. The hypoglycaemic event that occurs at approximately one day after monitoring began is coloured red and classified as very unusual. Figure 7 shows a trace with a few potentially unusual measurements for the first three days of monitoring. After day 3 a high proportion of the CGM measurements are classified as very unusual and are coloured red.
Discussion
Clinical CGM data and stochastic model generation
The aim of this study was to design a tool that could aid clinicians in identifying unusual CGM behaviour that should potentially be interpreted with care. Stochastic modelling methods from [31] and a method of colouring CGM traces were used to highlight unusual CGM behaviour clearly and efficiently, in either real-time or retrospectively.
Figures 1 and 2 give information about the raw data used to create the stochastic model. More than 99% of the data is within 2-10 mmol/L range, shown in Figure 2. There are several outliers in Figure 1 that have a very large change in glycaemia over the 5 minute measurement interval. The high data density means these outliers have little effect on the model fit, shown by the smooth and tight percentile lines in Figure 1. However, below 2 mmol/L there are 97 CGM measurements and due to the relatively low density of data the outliers have more impact on the model fit. This effect is clearly seen in the 95th percentile line of the model, which strays upward at levels below 2 mmol/L. Similarly, above 10 mmol/L there are only 232 measurements and the percentile lines all have a wave-like shape, again showing the effect of outliers where data density is low. A greater data density would alleviate these issues without changing the approach in this proof-of-concept.
The quality of data used to create a stochastic model will affect how future CGM measurements are classified. If a model is created using low quality data, containing a significant number of outliers, then the model could potentially classify future outlier measurements as ‘usual’ or ‘expected’. Data quality is particularly important when using small data sets, as single outliers can have more effect on the overall model. However, the growing use of databases and electronic records means that collecting large amounts of data that are fully representative will become easier, even for sub-cohorts. Thus, this work foreshadows an application with potential for good clinical use in future, as the method of creating such models is general.
Figure 3 shows the surface of the stochastic model. The colour gradient shows how the shape of the model changes in the domain of CGM n-1 and that a single, global probability density function is not applicable to this data set. Figure 4 further reinforces this with 5 pdf’s taken from the model at different CGM n-1 values, resulting in 5 different shaped density functions. These pdf’s are also used to show that the model fits the empirical data well. The model pdf’s (solid lines) overlay the empirical data (dotted lines) with only minor discrepancies.
Additionally, the model fit was checked using a 5-fold validation, with results shown in Table 1. Across the 25 MC runs, the 80% CI captured 83% of the data (median), the 90% CI captured 91% of the data (median) and 99% CI captured 99% of the data (median). These results show the model to capture the CGM data well over the expected range of CGM values, with little variation in the percentage of data captured in each CI.
Classification of representative CGM data
Figures 5, 6, 7 show 3 different CGM data sets and how the stochastic model classified the individual CGM measurements within them. Figure 5 shows a very stable, flat CGM trace with only small variations over the 3.5 days of monitoring. The CGM trace passes near all calibration measurements and there doesn’t appear to be any unusual CGM behaviour. The stochastic model classified almost the entire trace as dark blue indicating no unusual CGM behaviour. The interpretation of this trace would not likely be influenced with the additional information provided by the model.
Figure 6 shows a less stable CGM trace with a lot more variability. This trace contains a few yellow and red sections that potentially need to be interpreted with care. The focus of this discussion is the ‘hypoglycaemic event’ that occurs at ~ day 1. In the sequence of 5 measurements that lead up to the 1.8 mmol/L minimum, there are two drops of ~1 mmol/L per 5 minute measurement interval. The model has determined these are extreme outliers and consequently they have been coloured red. The trace then rises to above 4 mmol/L in 5 measurements, similarly with two rises of ~1 mmol/L per 5 minute measurement interval. Although the physiological limits of glucose rate-of-change are still unknown, the level of sensor error that has been reported in previous CGM studies [18, 22] suggest that this hypoglycaemic event could potentially be either glycaemia or sensor error.
Interestingly, the bottom of the ‘hypoglycaemic event’ at ~ day 1 contains a dark blue classification; indicating the measurement-to-measurement change immediately after the nadir was not unusual. This was not unexpected, because the lag 1 model classified the change between any two consecutive CGM measurements independent of previous classifications. Additionally, the data in Figure 1 is generally centred on the line CGM n-1 = CGM n and consequently the model surface in Figure 3 peaks along or near this line. Thus, for the hypoglycaemic event at day 1 when the CGM reported 1.8 mmol/L followed by 1.9 mmol/L, CGM n-1 ≈ CGM n , and the change was classified as expected (blue – within 80% CI).
It is important to note that the aim of the stochastic model is not to try and determine the accuracy of the CGM device or the specific cause of a drop in CGM glucose, but rather to highlight the fact it should be interpreted with care. Furthermore, if the stochastic model was implemented in a real-time clinical setting and the downward CGM measurements were observed, it would be beneficial for the clinician to know whether the sequence of measurements is typical of CGM devices and that patient cohort. It should also be noted that without an accurate BG measurement at ~1 day, no exact conclusion can be drawn about the whether the hypoglycaemic event in this data was a sensor artefact, or a true glycaemic event.
However, this lack of confirmation is often the reality with CGMs. Clinical protocols might use stochastic information to justify an added BG measurement to clarify a potentially significant event. After an event, such traces would yield insight not present at the bedside.
Figure 7 shows an example of CGM data that becomes increasingly more variable and unstable at approximately day 3 of monitoring. Before day 3, the CGM trace is predominantly blue and cyan with only small patches of yellow and occasionally red. However, after day 3 the CGM trace is almost entirely red indicating the stochastic model has classified these measurements as very unusual. The sudden apparent degradation of reliable CGM measurements could be due to a sensor failure. This is not an unreasonable hypothesis, given the sensors used in this study were validated for 3 days of continuous monitoring. Again, without more frequent, accurate BG measurements during the period after day 3 no definitive conclusions can be drawn. However, this example represents another potential use of this stochastic model classification method that might be useful to users of CGM devices.
Finally, the stochastic model and classification methods were used retrospectively in this study. For real-time use, a stochastic model would be generated using prior CGM data, and CGM measurements would be entered into the system in real-time. Classification of paired CGM measurements (CGM n-1 , CGM n ) takes a fraction of a second, so the corresponding colour coded segment of CGM trace would be displayed without significant delay (estimated less than 1 second). The major limitation in implementing the method in real-time is the ability to stream CGM data to a computer in real-time. Although the technology is available, no ‘off the shelf’ CGM devices currently offer this feature.
Conclusions
A stochastic model was shown to be capable of classifying CGM measurements to highlight unusual CGM behaviour. The method uses a colour coded CGM trace to convey the information quickly and efficiently and it is computationally light enough to be used retrospectively or in real-time.
There are several potential uses for the stochastic classification which include, but are not limited to, classification of hypoglycaemia and detection of potential sensor failure. Equally, they can augment alarm methods or be used to more optimally time BG measurements, such as in neonates where blood draws are restricted. Overall, while BG measurements are required to draw definitive conclusions about glycaemic events, the stochastic model provides another level of information to aid users in interpretation and decision making.
Financial support
UC Department of Mechanical Engineering, New Zealand, Health Research Council of New Zealand. The project described was supported in part by Grant Number RO1HD069622 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health.
References
Capes SE, Hunt D, Malmberg K, Gerstein HC: Stress hyperglycaemia and increased risk of death after myocardial infarction in patients with and without diabetes: a systematic overview. Lancet 2000, 355: 773–778. 10.1016/S0140-6736(99)08415-9
Finney SJ, Zekveld C, Elia A, Evans TW: Glucose control and mortality in critically ill patients. JAMA: the journal of the American Medical Association 2003, 290: 2041–2047. 10.1001/jama.290.15.2041
Krinsley JS: Association between hyperglycemia and increased hospital mortality in a heterogeneous population of critically ill patients. Mayo Clinic proceedings Mayo Clinic 2003, 78: 1471–1478.
McCowen KC, Malhotra A, Bistrian BR: Stress-induced hyperglycemia. Crit Care Clin 2001, 17: 107–124. 10.1016/S0749-0704(05)70154-8
Mizock BA: Alterations in fuel metabolism in critical illness: hyperglycaemia. Best Pract Res Clin Endocrinol Metab 2001, 15: 533–551. 10.1053/beem.2001.0168
Umpierrez GE, Isaacs SD, Bazargan N, You X, Thaler LM, Kitabchi AE: Hyperglycemia: an independent marker of in-hospital mortality in patients with undiagnosed diabetes. J Clin Endocrinol Metab 2002, 87: 978–982. 10.1210/jc.87.3.978
Van den Berghe G, Wouters P, Weekers F, Verwaest C, Bruyninckx F, Schetz M, Vlasselaers D, Ferdinande P, Lauwers P, Bouillon R: Intensive insulin therapy in the critically ill patients. N Engl J Med 2001, 345: 1359–1367. 10.1056/NEJMoa011300
Bistrian BR: Hyperglycemia and infection: which is the chicken and which is the egg? JPEN J Parenter Enteral Nutr 2001, 25: 180–181. 10.1177/0148607101025004180
Beardsall K, Vanhaesebrouck S, Ogilvy-Stuart AL, Vanhole C, Palmer CR, van Weissenbruch M, Midgley P, Thompson M, Thio M, Cornette L, et al.: Early Insulin Therapy in Very-Low-Birth-Weight Infants. N Engl J Med 2008, 359: 1873–1884. 10.1056/NEJMoa0803725
Le Compte A, Chase J, Lynn A, Hann C, Shaw G, Wong X, Lin J: Blood Glucose Controller for Neonatal Intensive Care: Virtual trials development and 1st clinical trials. Journal of Diabetes Science and Technology 2009, 3: 1066–1081.
Egi M, Bellomo R, Stachowski E, French CJ, Hart G: Variability of blood glucose concentration and short-term mortality in critically ill patients. Anesthesiology 2006, 105: 244–252. 10.1097/00000542-200608000-00006
Egi M, Bellomo R, Stachowski E, French CJ, Hart GK, Taori G, Hegarty C, Bailey M: Hypoglycemia and outcome in critically ill patients. Mayo Clinic proceedings Mayo Clinic 2010, 85: 217–224.
Hermanides J, Bosman RJ, Vriesendorp TM, Dotsch R, Rosendaal FR, Zandstra DF, Hoekstra JB, DeVries JH: Hypoglycemia is associated with intensive care unit mortality. Crit Care Med 2010, 38: 1430–1434. 10.1097/CCM.0b013e3181de562c
Krinsley JS: Glycemic variability: a strong independent predictor of mortality in critically ill patients. Crit Care Med 2008, 36: 3008–3013. 10.1097/CCM.0b013e31818b38d2
Carayon P, Gurses AP: A human factors engineering conceptual framework of nursing workload and patient safety in intensive care units. Intensive Crit Care Nurs 2005, 21: 284–301. 10.1016/j.iccn.2004.12.003
Chase JG, Andreassen S, Jensen K, Shaw GM: Impact of Human Factors on Clinical Protocol Performance: A Proposed Assessment Framework and Case Examples. Journal of Diabetes Science and Technology 2008, 2: 409–416.
Mackenzie I, Ingle S, Zaidi S, Buczaski S: Tight glycaemic control: a survey of intensive care practice in large English hospitals. Intensive Care Med 2005, 31: 1136. 10.1007/s00134-005-2677-2
Goldberg PA, Siegel MD, Russell RR, Sherwin RS, Halickman JI, Cooper DA, Dziura JD, Inzucchi SE: Experience with the continuous glucose monitoring system in a medical intensive care unit. Diabetes Technol Ther 2004, 6: 339–347. 10.1089/152091504774198034
Brunner R, Kitzberger R, Miehsler W, Herkner H, Madl C, Holzinger U: Accuracy and reliability of a subcutaneous continuous glucose-monitoring system in critically ill patients. Crit Care Med 2011, 39: 659–664. 10.1097/CCM.0b013e318206bf2e
Lorencio C, Leal Y, Bonet A, Bondia J, Palerm CC, Tache A, Sirvent JM, Vehi J: Real-Time Continuous Glucose Monitoring in an Intensive Care Unit: Better Accuracy in Patients with Septic Shock. Diabetes Technol Ther 2012, 14: 568–575.
De Block C, Manuel YKB, Van Gaal L, Rogiers P: Intensive insulin therapy in the intensive care unit: assessment by continuous glucose monitoring. Diabetes Care 2006, 29: 1750–1756. 10.2337/dc05-2353
Breton M, Kovatchev B: Analysis, modeling, and simulation of the accuracy of continuous glucose sensors. J Diabetes Sci Technol 2008, 2: 853–862.
Klonoff DC: Continuous Glucose Monitoring: Roadmap for 21st century diabetes therapy. Diabetes Care 2005, 28: 1231–1239. 10.2337/diacare.28.5.1231
Klonoff DC: A review of continuous glucose monitoring technology. Diabetes Technol Ther 2005, 7: 770–775. 10.1089/dia.2005.7.770
Chee F, Fernando T, van Heerden PV: Closed-loop glucose control in critically ill patients using continuous glucose monitoring system (CGMS) in real time. IEEE Trans Inf Technol Biomed 2003, 7: 43–53. 10.1109/TITB.2003.808509
Chee F, Fernando TL, Savkin AV, van Heeden V: Expert PID control system for blood glucose control in critically ill patients. IEEE Trans Inf Technol Biomed 2003, 7: 419–425. 10.1109/TITB.2003.821326
Clarke WL, Anderson S, Farhy L, Breton M, Gonder-Frederick L, Cox D, Kovatchev B: Evaluating the clinical accuracy of two continuous glucose sensors using continuous glucose-error grid analysis. Diabetes Care 2005, 28: 2412–2417. 10.2337/diacare.28.10.2412
Skyler JS: Continuous glucose monitoring: an overview of its development. Diabetes Technol Ther 2009, 11(Suppl 1):S5–10.
Cembrowski GS, Tran DV, Higgins TN: The use of serial patient blood gas, electrolyte and glucose results to derive biologic variation: a new tool to assess the acceptability of intensive care unit testing. Clinical chemistry and laboratory medicine : CCLM /FESCC 2010, 48: 1447–1454.
Watkinson PJ, Barber VS, Amira E, James T, Taylor R, Young JD: The effects of precision, haematocrit, pH and oxygen tension on point-of-care glucose measurement in critically ill patients: a prospective study. Ann Clin Biochem 2012, 49: 144–151. 10.1258/acb.2011.011162
Lin J, Lee D, Chase JG, Shaw GM, Le Compte A, Lotz T, Wong J, Lonergan T, Hann CE: Stochastic modelling of insulin sensitivity and adaptive glycemic control for critical care. Comput Meth Prog Bio 2008, 89: 141–152. 10.1016/j.cmpb.2007.04.006
Acknowledgements
The authors would like to acknowledge members of the CHYLD Study Group for collecting and compiling data for this study, and the CHYLD Study Steering Group for oversight and approval of the manuscript.
CHYLD Study Group: Steering Group: Jane Alsweiler; Geoff Chase; Jane Harding; Deborah Harris; Ben Thompson; Trecia Wouldes.
Group Members: Judith Ansell; Nicola Anstice; Jo Arthur; Coila Bevan; Kate Bluett; Ellen Campbell; Arijit Chakraborty; Tineke Crawford; Karen Frost; Greg Gamble; Rob Jacobs; Kelly Jones; Aaron Le Compte; Sapphire Martin; Gill Matheson; Grace McKnight; Christina McQuoid; Janine Paynter; Jenny Rogers; Matthew Signal; Heather Stewart; Anna Timmings; Rebecca Young; Sandy Yu
Author information
Authors and Affiliations
Corresponding author
Additional information
Competing interest
The authors declare that they have no competing interests.
Authors’ contributions
MS: Obtained main results and wrote the main body of the article, ALC: Assisted in obtaining results and writing, DLH: Collected clinical data and assisted in writing, PJW: Collected clinical data and assisted in writing, JEH: Collected clinical data and assisted in writing, JGC: Assisted in obtaining results and writing. All authors read and approved the final manuscript.
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
This article is published under license to BioMed Central Ltd. This is an Open Access article distributed under the terms of the Creative Commons Attribution License (http://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Signal, M., Le Compte, A., Harris, D.L. et al. Using Stochastic modelling to identify unusual continuous glucose monitor measurements and behaviour, in newborn infants. BioMed Eng OnLine 11, 45 (2012). https://doi.org/10.1186/1475-925X-11-45
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1475-925X-11-45