Abstract
Background
The global ppGpp-mediated stringent response in pathogenic bacteria plays an important role in the pathogenesis of bacterial infections. In Salmonella enterica serovar Typhimurium (S. Typhimurium), several genes, including virulence genes, are regulated by ppGpp when bacteria are under the stringent response. To understand the control of virulence genes by ppGpp in S. Typhimurium, agarose 2-dimensional electrophoresis (2-DE) combined with mass spectrometry was used and a comprehensive 2-DE reference map of amino acid-starved S. Typhimurium strain SH100, a derivative of ATCC 14028, was established.
Results
Of the 366 examined spots, 269 proteins were successfully identified. The comparative analysis of the wild-type and ppGpp0 mutant strains revealed 55 proteins, the expression patterns of which were affected by ppGpp. Using a mouse infection model, we further identified a novel virulence-associated factor, STM3169, from the ppGpp-regulated and Salmonella-specific proteins. In addition, Salmonella strains carrying mutations in the gene encoding STM3169 showed growth defects and impaired growth within macrophage-like RAW264.7 cells. Furthermore, we found that expression of stm3169 was controlled by ppGpp and SsrB, a response regulator of the two-component system located on Salmonella pathogenicity island 2.
Conclusions
A proteomic approach using a 2-DE reference map can prove a powerful tool for analyzing virulence factors and the regulatory network involved in Salmonella pathogenesis. Our results also provide evidence of a global response mediated by ppGpp in S. enterica.
Similar content being viewed by others
Background
The facultative intracellular bacterium Salmonella enterica causes a broad spectrum of diseases, such as gastroenteritis and bacteremia, which are typically acquired by oral ingestion of contaminated food or water. S. enterica serovar Typhimurium (S. Typhimurium) causes enterocolitis in humans and a typhoid-like systemic infection in mice.
Several virulence genes associated with Salmonella pathogenicity islands (SPIs) and the virulence plasmid have been characterized in S. Typhimurium. Two type III secretion systems (T3SS) encoded by SPI-1 and SPI-2 play central roles in Salmonella pathogenesis. SPI-1 is essential for the invasion of host cells and the induction of apoptosis in infected macrophages [1, 2]. SPI-2 T3SS primarily confers survival and replication on macrophages and is required for systemic infection in the mouse infection model [3, 4]. Expression of SPI-2 genes is induced within a modified phagosome, called the Salmonella-containing vacuole (SCV), in infected macrophages [5]. Induction of SPI-2 genes depends on a two-component regulatory system, SsrA/SsrB, encoded within the SPI-2 region [6]. Expression of SsrAB is also mediated by two-component regulatory systems, OmpR/EnvZ and PhoP/PhoQ, which sense osmotic stress and cation limitation, respectively [7, 8]. In addition, a global transcriptional regulator, SlyA, which interacts directly with the ssrA promoter region, is involved in the expression of SPI-2 T3SS [9–11].
During infection of mammalian hosts, S. Typhimurium has to rapidly adapt to different environmental conditions encountered in its passage through the gastrointestinal tract and its subsequent uptake into epithelial cells and macrophages. Thus, establishment of infection within a host requires coordinated expression of a large number of virulence genes necessary for the adaptation between extracellular and intracellular phases of infection. It has been demonstrated that the stringent response plays an important role in the expression of Salmonella virulence genes during infection [12–14].
The stringent response is mediated by the signal molecules, guanosine tetraphosphate (ppGpp) and guanosine pentaphosphate (pppGpp) (both are referred to as ppGpp in this manuscript), which accumulate in bacterial cells and exert both positive and negative effects on the transcription of many genes. ppGpp plays an important role in the virulence of pathogenic bacteria [15]. In Gram-negative bacteria, ppGpp is synthesized by two tynthases, the synthase I and the synthase II, which are encoded by the relA and spoT genes, respectively [16]. These enzymes respond differently to environmental conditions. RelA is activated by the binding of uncharged tRNA to ribosomes upon amino acid starvation. SpoT is induced during the exponential growth phase and responds to other changes in environmental conditions, specifically a lack of carbon sources or energy deprivation [17]. ppGpp binds directly to the β and β' subunits of RNA polymerase (RNAP), leading to destabilization of the RNAP-rRNA promoter open complex [18]. Moreover, the stringent response is increased by the availability of free RNAP, which gives rise to σ competition [19]. ppGpp indirectly activates the expression of many stress-induced genes by its release from RNAP σ70-dependent promoters and by facilitating the use of alternativeσ factors. It has been shown that ppGpp is not only essential for surviving periods of stress but also for the interaction of bacteria with their host [20].
In case of S. Typhimurium, a mutant strain deficient in both relA and spoT (ΔrelA ΔspoT) shows marked reductions in both bacterial invasion into host cells and proliferation in macrophages [12, 13]. Furthermore, the virulence of the ΔrelA ΔspoT mutant is severely attenuated in mice [12, 13]. ppGpp controls the expression of SPI-1 to -5 and Spv through their transcriptional regulators HilA, InvF, RtsA, SsrA, SlyA, and SpvR [12–14, 21]. These observations indicate that ppGpp may play a major role in Salmonella virulence via the altered expression of regulatory genes. Because ppGpp has been shown to affect the expression of many virulence genes in S. Typhimurium, it is likely that there are additional virulence genes among the ppGpp-regulated genes.
In this study, we constructed an agarose 2-dimensional electrophoresis (2-DE) reference map of S. Typhimurium grown under amino acid starvation to identify ppGpp-regulated proteins from whole-cell preparations. By comparative proteomic analysis of ppGpp-regulated and Salmonella- specific proteins, we identified a novel virulence factor, STM3169, required for intracellular survival within macrophages.
Results and Discussion
Agarose 2-DE reference map of S. Typhimurium with induced stringent responses
Because the correlation between mRNA and protein expression levels is nonpredictive, the direct measurement of protein expression is essential for the analysis of biological processes [22]. 2-DE allows several hundred proteins to be displayed on a single gel, thus producing a direct and global view of the proteome at a given time point [23]. Agarose 2-DE takes advantage of the process of protein separation over a broad range [24, 25]. In this study, to separate and identify more proteins, we applied agarose 2-DE to the bacterial proteome, and also used 12% and 15% SDS-PAGE gels for the second dimension. Whole-cell proteins were obtained from the S. Typhimurium strain SH100, a derivative of ATCC 14028, with the stringent response induced by serine hydroxamate, as described previously [26]. Agarose 2-DE was performed at least three times on independent samples. More than 350 protein spots from the strain were detected on each 2-DE gel stained with Coomassie Brilliant Blue. To identify proteins on the agarose 2-DE gels, we excised 230 spots from the 12% gel and 136 spots from the 15% gel. We finally identified a total of 360 proteins (273 proteins from the 12% gel [Figure 1A] and 87 proteins from the 15% gel [Figure 1B]) by MS/MS analysis out of 307 protein spots (232 spots from the 12% gel and 75 spots from the 15% gel) that were successfully excised (see additional file: 1). In total, 267 proteins were obtained from the gels, with 40 proteins identified as being redundant. The highest and lowest molecular masses of identified proteins were 93.4 kDa for AcnB (aconitate hydrase 2, spot 188) and 7.4 kDa for CspC (cold-shock protein, spot 303), respectively. Fifty spots (35 spots from the 12% gel and 15 spots from the 15% gel) were found in a basic range. Interestingly, 78 protein spots (25.4%) were annotated as putative proteins on the genome of the S. Typhimurium LT2 strain, which is more than 98% identical in sequence to the 14028 strain [27].
Agarose 2-DE reference map of the S . Typhimurium strain SH100, prepared using a 12% gel focused on high-molecular-mass proteins (A) and a 15% gel focused on low-molecular-mass proteins (B). Strain SH100 was grown under amino acid starvation as described previously [26]. Gels are stained with Coomassie Brilliant Blue. Identified spots are numbered (corresponding to the spot numbers in additional file: 1. Proteins identified on the reference map).
We estimated the molecular weight of the protein spots on the 2-DE gels and compared them with the theoretical molecular weight of strain SH100. While most of the estimated molecular weight values matched the theoretical values, we found 14 protein spots on the map that had different experimental and predicted molecular weights values (Figure 2). These proteins might be post-translationally modified by proteolytic processing, phosporylatoin of multiple amino acid residues and/or an artifact caused by sample preparation. For example, the experimental molecular weight of OmpA indicated that the protein was likely processed by a proteolytic enzyme, because two different spots (spot nos. 152 and 287) were identified as OmpA, the experimental masses of which were significantly lower than the theoretical values. Similar results were described in other reports [28, 29].
Comparison of the gel-estimated and theoretically calculated molecular weight (Mw) of the identified protein spots. Arrows indicate spot numbers and identified proteins, which have different Mw between the experimental and the predicted ones. The proteins in the area enclosed by the dotted lines denote that they have an experimental Mw within ± 25% of the predicted molecular mass.
Next, we classified proteins identified on the map using the KEGG pathway database. While 156 proteins (45.3%) were classified into several metabolic categories (carbohydrate, energy, lipid, nucleotide, amino acid, and other amino acids), 70 proteins (22.8%) were grouped in the no entry category, which means that these proteins do not belong to the other categories. This category contained 20 known virulence-associated proteins, including flagella and flagella biosynthesis proteins (FliC, FljB, FliY, FliG, FliM, and FliD), SPI-1 effectors (SipD, SopB, and SopE2), an SPI-1 translocase (SipC), an iron transporter (SitA), superoxide dismutases (SodA, SodB, SodC1, and SodC2), a quorum-sensing protein (LuxS), a two-component response regulator (PhoP), peptidyl-prolyl cis-trans isomerases (FkpA and SurA), and a periplasmic disulfide isomerase (DsbA).
Identification of ppGpp-regulated proteins using comparative proteomics
To identify proteins associated with the stringent response in S. Typhimurium, we compared the agarose 2-DE pattern for each total protein prepared from amino acid-starved S. Typhimurium SH100 and ΔrelA ΔspoT strain (TM157) (Figure 3). As shown in Table 1, 24 protein spots (23 proteins) were found at higher levels in SH100 than in TM157, while 23 protein spots were found at lower levels in SH100 than in TM157. We focused on 23 proteins, which were positively regulated by ppGpp in the stringent response.
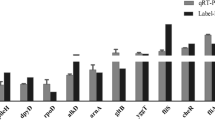
Of these proteins, six genes (treA, ugpB, ynhG, yliB, ugpB, degQ) had previously been identified as ppGpp-regulated genes in E. coli at the transcriptional level [30]. In S. Typhimurium, it has been shown that ppGpp controls the expression of known virulence-associated genes, including sipC, fliY, sopB, and sodC1, in response to growth conditions relevant to host infection [14]. Thus, to confirm the results from the comparative proteomic analysis, mRNA levels of the remaining 13 genes were assessed by qRT-PCR. As a result, mRNA expression levels of eight genes (stm3169, cpdB, tolB, ydgH, oppA, yajQ, yhbN, ytfJ) were significantly higher in SH100 than in TM157 under stringent conditions (Table 1).
Identification of novel virulence-associated factors regulated by ppGpp
Among 13 genes newly identified as ppGpp regulated, 12 genes were present in non-pathogenic E. coli K-12 strain. Therefore, to examine whether ppGpp-regulated putative or hypothetical proteins could contribute to the virulence of S. Typhimurium, we chose Salmonella-specific protein, STM3169, which is present in S. Typhimurium, but is absent in the E. coli K-12 strain (Figure 4[27, 31]). To determine the roles of STM3169 in virulence, a deletion mutant was constructed in the S. Typhimurium wild-type SH100 strain, and its virulence was assessed by a mouse mixed infection using a competitive index analysis. As shown in Figure 5A, mouse mixed infections showed that disruption of the stm3169 gene conferred a defect in virulence in mice, and that successful complementation was achieved for TH973 (Δstm3169::kan) by expression of intact STM3169 from a plasmid. These findings provide the first evidence that STM3169 functions as a virulence factor of S. Typhimurium in a mouse infection model.
The S . Typhimurium-specific protein STM3169 is regulated by ppGpp in the stringent response. (A) Comparison of the STM3169 protein expression in the wild-type SH100 and ΔrelA ΔspoT strain (TM157). An asterisk indicates that the difference was statistically significant (P < 0.05). (B) Genetic map of genes (open arrows) coding STM3169 within Salmonella-specific locus (gray arrows) and genes flanking the locus (closed arrows).
STM3169 is a novel virulence protein. Competitive index was determined at 48 h after infection in the spleen (A). Effects of stm3169 disruption on the invasion (B) and the intracellular survival (C) in the mouse macrophage cell lines, RAW264.7. Cells treated with IFN-γ were infected with S. Typhimurium wild-type and the mutant strains at a multiplicity of infection of 1. At 2 h and 24 h after infection, macrophages were lysed and the bacterial number was determined. Asterisks indicate that differences were statistically significant (P < 0.05).
Because it is believed that intracellular Salmonella is likely to be restricted to the acquisition of nutrient substrates from infected host cells, the stringent response could occur in SCV. Thus, we next analyzed the contribution of STM3169 to intracellular survival of S. Typhimurium in macrophages. In accordance with previous data that a ppGpp0 mutant strain deficient in both spoT and relA genes resulted in a severe reduction of intracellular proliferation and suvival [12]. In contrast to the wild-type level of invasion, intracellular survival of TH973 in RAW264.7 cells was reduced, compared with that of the wild-type strain. The reduced CFU of TH937 in IFN-γ treated-RAW264.7 cells was not more severe than that in the ΔrelA ΔspoT double mutant, ΔssaV (SH113, SPI-2 T3SS component-defected mutant), and ΔssrB (YY1, SPI-2 regulator mutant) strain, but was equal to that in the ΔsseF (TM548, SPI-2 effector mutant) strain (Figure 5B and 5C). These results suggest that the expression of additional virulence factors, like STM3169, in macrophages might be affected in a highly avirulent phenotype of a ppGpp-deficient strain in mice.
stm3169 is regulated by the SPI-2 transcriptional regulator ssrB
It has been demonstrated that ppGpp mediates the expression of virulence-associated genes involved in bacterial invasion and intracellular growth and survival via global and/or gene-specific transcriptional regulators in S. Typhimurium [12, 14]. Since intracellular growth and suvival of Salmonella in macrophages is dependent upon SPI-2 function, we next confirmed whether expression of stm3169 is regulated by the SsrAB two-component system, which positively controls the expression of SPI-2 genes as well as other genes belonging to the SsrB regulon [32]. To test this, we constructed S. Typhimurium strains carrying stm3169::lacZ transcriptional fusions on the chromosome in the wild-type (SH100) and ΔrelA ΔspoT (TM157) genetic background. Salmonella strains carrying the stm3169::lacZ fusion gene (TH1162 and TH1164) were grown in defined MgM medium (pH 5.8) with 0.1% casamino acids and measured β-galactosidase activity. The transcription levels of stm3169::lacZ fusion were significantly decreased in TM157 (Figure 6A). The reduced level was restored to the wild-type level by the introduction of an arabinose-inducible plasmid expressing His6-tagged RelA protein. We next transduced the ssrB mutation (ΔssrB::cat) into a stm3169::lacZ fusion strain (TH1162). Strains carrying the stm3169::lacZ fusion gene with the ssrB mutation were grown in MgM medium (pH 5.8), and β-galactosidase activity was measured. Control experiments were performed with the ssaG::lacZ fusion gene (TM129). ssaG expression is strongly controlled by SsrB [33]. Similar to ssaG::lacZ, the transcription level of the stm3169::lacZ fusion gene was significantly decreased in strains carrying the ssrB mutation (Figure 6B). Complementation was partially achieved for TM423 by expression of SsrB (SsrB-FLAG) on a plasmid (Figure 6B), probably due to the constitutive expression of SsrB from multi-copy-number palsmid pFLAG-CTC. Collectively, these data suggest that the novel virulence-associated factor STM3169 was regulated by the SPI-2 two-component regulatory system SsrAB as well as by ppGpp.
STM3169 is regulated by ppGpp and ssrB. Transcriptional activity of stm3169 in Salmonella muntant strains. Salmonella Δrel A ΔspoT (A), ΔssrB (B), and ΔrelA ΔspoT ΔssrB (C) mutant strains carrying stm3196::lacZ fusion were incubated in MgM medium (pH5.8) for 18 h. The promoter activity of stm3169 was estimated by mesuring the β-garactosidase activity. L-arabinose (a final concentration of 0.001%) and IPTG (a final concentration of 0.01 mM) were added in the medium for induction of RelA on pRelA and for SsrB on pSsrB, respectively. Asterisks indicate that differences were statistically significant (P < 0.05).
It has been reported that ppGpp regulates SPI-2-encoded genes under aerobic condition [14]. To further characterize the transcriptional regulation of stm3169 by ppGpp and SsrB, we constructed a ΔrelA ΔspoT ΔssrB triple mutant strain (YY2), and examined the affect of the transcriptional activity on stm3169::lacZ fusion gene. While the transcriptional activity of stm3169::lacZ fusion in the triple mutant strain was significantly reduced at the same level of ΔrelA ΔspoT double mutant strain, it could be restored by introduction of plasmid pSsrB expressing SsrB-FLAG but not pRelA expressing His6-tagged RelA (Figure 6C). These results indicate that ppGpp is controlled the expression of stm3169 through SsrB.
STM3169 is homologous to DctP in Rhodobacter capsulatus with a 31% identity and a 73% similarity. DctP, along with DctQ and DctM, constitutes a tripartite ATP-independent periplasmic transporter (TRAP-T) system involved in succinate utilization, and DctP plays a role as an extracytoplasmic solute receptor in this transporter [34]. STM3170 and STM3171, which are located immediately downstream from STM3169, have a 66% and an 80% similarity with DctQ and DctM, respectively. These suggest that the TRAP-T in S. Typhimurium is composed of stm3169, stm3170, and stm3171 genes. In addition, two hypothetical operons, yiaOMN and stm4052-4054, are annotated as TRAP-T in the S. Typhimurium strain LT2 [31]. Recently, it has been reported that the TRAP-T (SiaPQM) in Haemophilus influenzae is essential for LPS sialylation and virulence [35]. Further research is necessary to determine the role of these transporters in S. Typhimurium virulence.
Conclusions
We constructed an agarose 2-DE reference map of amino-acid starved S. Typhimurium and identified a novel virulence-associated factor, STM3169, regulated by ppGpp by applying the map to comparative proteomics. stm3169 is also regulated by an SPI-2 two-component regulator, SsrB. Recently, it has been reported that the lack of ppGpp synthesis in Salmonella strains attenuates virulence and induces immune responses in mice [36]. Thus, further analysis of proteins regulated by ppGpp may lead to the development of new vaccines.
Methods
Bacterial strains, primers, and culture conditions
The bacterial strains and plasmids used in this study are listed in Table 2. The oligonucleotide primers used are listed in Table 3. Bacteria were grown in Luria-Bertani (LB) medium or on LB agar under conditions suitable for selection for resistance to ampicillin (100 μg/mL), chloramphenicol (25 μg/mL), nalidixic acid (50 μg/mL), or spectinomycin (50 μg/mL), as appropriate. To induce the bacterial stringent response, serine hydroxamate (Sigma; 0.005%), an inhibitor of serine tRNA synthetase, was added to a 12 h culture in LB broth, and the bacteria were further incubated for 1 h [26]. Magnesium minimal medium (MgM, pH 5.8) was used to induce SPI-2 gene expression [6].
Construction of mutants
Nonpolar mutants of relA and spoT were constructed by allele exchange using the temperature- and sucrose-sensitive suicide vector pCACTUS [37]. The relA and spoT genes were amplified by PCR with the following primers: (1) relA-FW and relA-RV for relA and (2) spoT-FW and spoT-RV for spoT. S. Typhimurium strain SH100 genomic DNA was used as the template. The PCR products were cloned into TA cloning vector pGEM-T Easy (Promega) generating plasmid pGEM-relA and pGEM-spoT, respectively. A disruption mutation of relA was created by the insertion of the HincII-digested promoterless cat gene into a unique NruI site in the coding sequence of relA on pGEM-relA. The resulting plasmid pGEM-relA::cat was digested with BglII and then self-ligated, yielding plasmid pGEM-ΔrelA::cat. In contrast, the spoT gene was disrupted by the insertion of a SmaI-digested Kmr-encoding gene (kan) cassette from pUC18K [38] into NruI sites in the coding sequence of spoT on pGEM-spoT, thus generating pGEM-ΔspoT::kan. The disrupted gene was then subcloned using SalI and SphI into similarly digested pCACTUS, and the resulting plasmid was introduced into strain SH100 by electroporation for allele exchange mutagenesis, which was carried out as described previously [39]. ΔrelA ΔspoT mutant strain was created by phage P22-mediated transduction [40].
The PCR-based λ Red recombinase system using pKD46 and pKD4 was performed to disrupt stm3169 or sseF[41]. The growth rate of these mutant strains in LB and MgM (pH5.8) broth showed the same levels to wild-type strain.
To construct ΔrelA ΔspoT ΔssrB mutant strain, the cloned ssrB gene was disrupted by the insertion of a Tetr-encoding gene (tet) cassette, which was amplified with pAC-tet-FW and pAC-tet-RV primers using pACYC184 (New England Biolabs) as template. The ΔssrB::tet fragment was amplified by PCR using ssrB-FW and ssrB-RV primers, and the resulting PCR product was introduced into S. Typhimurium SH100 carrying pKD46. The disrupted genes were transferred by phage P22 transduction into ΔrelA ΔspoT mutant strain TM157.
To construct ssaG::lacZ and stm3169::lacZ transcriptional fusions, pLD-ssaGZ and pLD-stm3169Z were transferred from Escherichia coli SM10λpir to S. Typhimurium SH100 by conjugation. The fusions were introduced into SH100, ΔrelA ΔspoT (TM157), ΔssrB::tet (YY3), and ΔssaV (SH113) mutant strains by phage P22-mediated transduction. All constructs were verified by PCR or DNA sequencing.
Construction of plasmids
For construction of the complementing plasmid, pMW-Stm3169, stm3169 gene was amplified by PCR with stm3169-FW and stm3169-RV primers. S. Typhimurium SH100 genomic DNA was used as the template. The PCR products were digested with BglII and XhoI, and cloned into the Bglll-XhoI site on pMW118 (Nippon Gene), generating plasmid pMW-Stm3169.
To construct pRelA and pSsrB, the target genes were amplified by PCR with the following primers: relA-FW2 and relA-RV2 for relA and ssrB-FW and ssrB-RV for ssrB. The PCR product containing relA was digested with XhoI-HindIII and cloned into the same sites on pBAD-HisA (Invitrogen). The PCR product containing ssrB was digested with XhoI-BamHI and cloned into the same sites on pFLAG-CTC (Sigma). pRelA and pSsrB expressed His6-tagged RelA and SsrB-FLAG fusion protein, respectively.
To construct lacZ transcriptional fusions, the DNA fragments containing (predicted) promoter regions of ssaG were amplified by PCR using the primers ssaG-Pro-FW and ssaG-Pro-RV, and those containing promoter regions of stm3169 were amplified using stm3169-Pro-FW and stm3169-Pro-RV. The PCR products digested with SalI and BamHI were ligated into the same sites of pLD-lacZ Ω [39].
Sample preparation for agarose 2-DE
Agarose 2-DE samples were prepared from amino-acid starved S. Typhimurium strain SH100, as well as relA and spoT double knockout strain TM157 (ΔrelA ΔspoT). The cell pellets were washed twice with cold phosphate-buffered saline (PBS) and dissolved in lysis buffer containing 5 M urea, 1 M thiourea, 0.05% w/v β-mercaptoethanol, and one tablet of protein inhibitor (Complete Mini EDTA-free; Roche Diagnostics, Mannheim, Germany), which was dissolved in 10 mL of the solution. The lysates were centrifuged (104,000 × g, 20 min, 4°C) and the clear supernatant was used.
Proteome analysis
We performed proteome analysis according to the procedures of Oh-Ishi et al.[25] and Kuruma et al.[42]. An aliquot of 200-300 μL (containing 500 μg of protein) of sample solution was subjected to first-dimension IEF at 667 V for 18 h at 4°C, followed by second-dimension SDS-PAGE. The slab gel was stained with CBB R-350 (PhastGel Blue R; GE Healthcare).
Protein spots were excised from a destained gel with 50% (v/v) ACN and dried under vacuum. The proteins were digested in the gel with trypsin. Digested fragments of 15 pmol were loaded on a Liquid Chromatography-Mass Spectrometry/Mass Spectrometry (LC-MS/MS), which consisted of Nanospace SI-2 (Shiseido Fine Chemicals), an HPLC (LCQ Deca), and an ion trap mass spectrometer (Thermo Finnigan). We identified a protein from measured masses of the tryptic peptides and their MS/MS fragments using the SEQUEST program. When the top-ranked candidates had SEQUEST scores lower than 90, we inspected the raw MS and MS/MS spectra of peptides to judge their qualities. We classified identified proteins according to the Kyoto Encyclopedia of Genes and Genomes (KEGG) PATHWAY database http://www.genome.ad.jp/kegg/pathway.html.
Gel-to-gel comparisons between SH100 and TM153 were performed for two separately prepared samples. Each scanned 2-DE gel image was analyzed with the gel image analysis software SameSpots (Progenesis).
RNA extraction and quantitative real-time PCR
S. Typhimurium strains were grown in LB and ppGpp expression was induced as described above. Total RNA was isolated from the bacterial culture using RNAprotect Bacteria Reagent and the RNeasy Protect Bacteria Mini Kit with the gDNA Eliminator spin column (Qiagen) according to the manufacturer's instructions. cDNA was synthesized using the QuantiTect Reverse Transcription Kit (Qiagen). Real-time PCR was performed with the primer pairs listed in Table 3 using QuantiTect SYBR Green and the 7900HT Sequence Detection System (Applied Biosystems). The data were analyzed using the comparative Ct method (Applied Biosystems). Transcription of the target gene was normalized to the levels of gyrA mRNA.
Mouse infections
For the competitive index assay, female BALB/c mice (5-6 weeks old) were used for the mouse infection study and were housed at Kitasato University according to the standard Laboratory Animal Care Advisory Committee guidelines. Mice were inoculated by intraperitoneal infection with 100 μL of inoculum containing a total of 1 × 105 bacteria (each strain at 5 × 104), consisting of an equal number of wild-type and mutant strains. At 48 h after infection, the mice were sacrificed by carbon dioxide inhalation. The spleens were homogenized in cold PBS by mechanical disruption. The number of each strain in the spleen was determined by plating a dilution series of the lysate onto LB agar alone and LB agar with appropriate antibiotics. Each competitive index value was calculated as [mutant/wild-type] output/[mutant/wild-type] input and represented as the mean of at least three independent infections.
Macrophage survival assay
Cells of a mouse macrophage-like line, RAW264.7, were diluted in DMEM containing 10% FBS and seeded in 24-well plates at a density of 5 × 105 cells per well. S. Typhimurium strains were used to infect RAW264.7 cells at a multiplicity of infection of 1. The bacteria were centrifuged onto the cells (500 ×g, 5 min) and incubated for 25 min at 37°C in a 5% CO2 incubator. Cells were washed three times with PBS, and DMEM containing interferon-γ (IFN-γ) (100 units/well; Peprotech) and gentamicin (100 μg/mL; Sigma) was added. After 95 min of incubation, the medium was replaced with DMEM containing IFN-γ (100 units/well) and gentamicin (10 μg/mL). The number of intracellular bacteria was determined at 2 h and 24 h after infection. For the enumeration of intracellular bacteria, the cells were washed three times with PBS and lysed in 1% Triton X-100, and bacteria were quantified by spreading serial 10-fold dilutions of RAW264.7 cell lysates on LB agar plates to count the colony-forming units (CFU). Each experiment was repeated three times.
β-galactosidase assay
β-galactosidase activities of reporter gene fusions were determined according to a standard procedure [43].
Statistical analysis
The competitive index, mRNA expression, and bacterial proliferation in macrophage cells were compared using Student's t-test. For comparative proteomics, the intensity of the spot was compared by one-way ANOVA. Values of P < 0.05 were considered statistically significant.
References
Galan JE, Curtiss R: Cloning and molecular characterization of genes whose products allow Salmonella typhimurium to penetrate tissue culture cells. Proc Natl Acad Sci USA. 1989, 86 (16): 6383-6387. 10.1073/pnas.86.16.6383.
Lundberg U, Vinatzer U, Berdnik D, von Gabain A, Baccarini M: Growth phase-regulated induction of Salmonella-induced macrophage apoptosis correlates with transient expression of SPI-1 genes. J Bacteriol. 1999, 181 (11): 3433-3437.
Shea JE, Hensel M, Gleeson C, Holden DW: Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996, 93 (6): 2593-2597. 10.1073/pnas.93.6.2593.
Ochman H, Soncini FC, Solomon F, Groisman EA: Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci USA. 1996, 93 (15): 7800-7804. 10.1073/pnas.93.15.7800.
Steele-Mortimer O, Brumell JH, Knodler LA, Meresse S, Lopez A, Finlay BB: The invasion-associated type III secretion system of Salmonella enterica serovar Typhimurium is necessary for intracellular proliferation and vacuole biogenesis in epithelial cells. Cell Microbiol. 2002, 4 (1): 43-54. 10.1046/j.1462-5822.2002.00170.x.
Beuzon CR, Banks G, Deiwick J, Hensel M, Holden DW: pH-dependent secretion of SseB, a product of the SPI-2 type III secretion system of Salmonella typhimurium. Mol Microbiol. 1999, 33 (4): 806-816. 10.1046/j.1365-2958.1999.01527.x.
Bijlsma JJ, Groisman EA: The PhoP/PhoQ system controls the intramacrophage type three secretion system of Salmonella enterica. Mol Microbiol. 2005, 57 (1): 85-96. 10.1111/j.1365-2958.2005.04668.x.
Lee AK, Detweiler CS, Falkow S: OmpR regulates the two-component system SsrA-ssrB in Salmonella pathogenicity island 2. J Bacteriol. 2000, 182 (3): 771-781. 10.1128/JB.182.3.771-781.2000.
Linehan SA, Rytkonen A, Yu XJ, Liu M, Holden DW: SlyA regulates function of Salmonella pathogenicity island 2 (SPI-2) and expression of SPI-2-associated genes. Infect Immun. 2005, 73 (7): 4354-4362. 10.1128/IAI.73.7.4354-4362.2005.
Navarre WW, Halsey TA, Walthers D, Frye J, McClelland M, Potter JL, Kenney LJ, Gunn JS, Fang FC, Libby SJ: Co-regulation of Salmonella enterica genes required for virulence and resistance to antimicrobial peptides by SlyA and PhoP/PhoQ. Mol Microbiol. 2005, 56 (2): 492-508. 10.1111/j.1365-2958.2005.04553.x.
Okada N, Oi Y, Takeda-Shitaka M, Kanou K, Umeyama H, Haneda T, Miki T, Hosoya S, Danbara H: Identification of amino acid residues of Salmonella SlyA that are critical for transcriptional regulation. Microbiology. 2007, 153 (Pt 2): 548-560. 10.1099/mic.0.29259-0.
Pizarro-Cerda J, Tedin K: The bacterial signal molecule, ppGpp, regulates Salmonella virulence gene expression. Mol Microbiol. 2004, 52 (6): 1827-1844. 10.1111/j.1365-2958.2004.04122.x.
Song M, Kim HJ, Kim EY, Shin M, Lee HC, Hong Y, Rhee JH, Yoon H, Ryu S, Lim S, et al: ppGpp-dependent stationary phase induction of genes on Salmonella pathogenicity island 1. J Biol Chem. 2004, 279 (33): 34183-34190. 10.1074/jbc.M313491200.
Thompson A, Rolfe MD, Lucchini S, Schwerk P, Hinton JC, Tedin K: The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J Biol Chem. 2006, 281 (40): 30112-30121. 10.1074/jbc.M605616200.
Dalebroux ZD, Svensson SL, Gaynor EC, Swanson MS: ppGpp conjures bacterial virulence. Microbiol Mol Biol Rev. 2010, 74 (2): 171-199. 10.1128/MMBR.00046-09.
Cashel M, Gentry DR, Hernandez DR, Vinella D: The stringent response. Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology. 1996, 2
Magnusson LU, Farewell A, Nystrom T: ppGpp: a global regulator in Escherichia coli. Trends Microbiol. 2005, 13 (5): 236-242. 10.1016/j.tim.2005.03.008.
Paul BJ, Ross W, Gaal T, Gourse RL: rRNA transcription in Escherichia coli. Annu Rev Genet. 2004, 38: 749-770. 10.1146/annurev.genet.38.072902.091347.
Jishage M, Kvint K, Shingler V, Nystrom T: Regulation of sigma factor competition by the alarmone ppGpp. Genes Dev. 2002, 16 (10): 1260-1270. 10.1101/gad.227902.
Braeken K, Moris M, Daniels R, Vanderleyden J, Michiels J: New horizons for (p)ppGpp in bacterial and plant physiology. Trends Microbiol. 2006, 14 (1): 45-54. 10.1016/j.tim.2005.11.006.
Zhao G, Weatherspoon N, Kong W, Curtiss R, Shi Y: A dual-signal regulatory circuit activates transcription of a set of divergent operons in Salmonella typhimurium. Proc Natl Acad Sci USA. 2008, 105 (52): 20924-20929. 10.1073/pnas.0807071106.
Chevalier F: Highlights on the capacities of "Gel-based" proteomics. Proteome Sci. 2010, 8: 23-10.1186/1477-5956-8-23.
Bae SH, Harris AG, Hains PG, Chen H, Garfin DE, Hazell SL, Paik YK, Walsh BJ, Cordwell SJ: Strategies for the enrichment and identification of basic proteins in proteome projects. Proteomics. 2003, 3 (5): 569-579. 10.1002/pmic.200300392.
Oh-Ishi M, Maeda T: Disease proteomics of high-molecular-mass proteins by two-dimensional gel electrophoresis with agarose gels in the first dimension (Agarose 2-DE). J Chromatogr B Analyt Technol Biomed Life Sci. 2007, 849 (1-2): 211-222. 10.1016/j.jchromb.2006.10.064.
Oh-Ishi M, Satoh M, Maeda T: Preparative two-dimensional gel electrophoresis with agarose gels in the first dimension for high molecular mass proteins. Electrophoresis. 2000, 21 (9): 1653-1669. 10.1002/(SICI)1522-2683(20000501)21:9<1653::AID-ELPS1653>3.0.CO;2-9.
Tosa T, Pizer LI: Effect of serine hydroxamate on the growth of Escherichia coli. J Bacteriol. 1971, 106 (3): 966-971.
Jarvik T, Smillie C, Groisman EA, Ochman H: Short-term signatures of evolutionary change in the Salmonella enterica serovar Typhimurium 14028 genome. J Bacteriol. 2010, 192 (2): 560-567. 10.1128/JB.01233-09.
Jing HB, Yuan J, Wang J, Yuan Y, Zhu L, Liu XK, Zheng YL, Wei KH, Zhang XM, Geng HR, et al: Proteome analysis of Streptococcus suis serotype 2. Proteomics. 2008, 8 (2): 333-349. 10.1002/pmic.200600930.
Ying T, Wang H, Li M, Wang J, Shi Z, Feng E, Liu X, Su G, Wei K, Zhang X, et al: Immunoproteomics of outer membrane proteins and extracellular proteins of Shigella flexneri 2a 2457T. Proteomics. 2005, 5 (18): 4777-4793. 10.1002/pmic.200401326.
Traxler MF, Summers SM, Nguyen HT, Zacharia VM, Hightower GA, Smith JT, Conway T: The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol Microbiol. 2008, 68 (5): 1128-1148. 10.1111/j.1365-2958.2008.06229.x.
McClelland M, Sanderson KE, Spieth J, Clifton SW, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al: Complete genome sequence of Salmonella enterica serovar Typhimurium LT2. Nature. 2001, 413 (6858): 852-856. 10.1038/35101614.
Worley MJ, Ching KH, Heffron F: Salmonella SsrB activates a global regulon of horizontally acquired genes. Mol Microbiol. 2000, 36 (3): 749-761. 10.1046/j.1365-2958.2000.01902.x.
Walthers D, Carroll RK, Navarre WW, Libby SJ, Fang FC, Kenney LJ: The response regulator SsrB activates expression of diverse Salmonella pathogenicity island 2 promoters and counters silencing by the nucleoid-associated protein H-NS. Mol Microbiol. 2007, 65 (2): 477-493. 10.1111/j.1365-2958.2007.05800.x.
Kelly DJ, Thomas GH: The tripartite ATP-independent periplasmic (TRAP) transporters of bacteria and archaea. FEMS Microbiol Rev. 2001, 25 (4): 405-424. 10.1111/j.1574-6976.2001.tb00584.x.
Jenkins GA, Figueira M, Kumar GA, Sweetman WA, Makepeace K, Pelton SI, Moxon R, Hood DW: Sialic acid mediated transcriptional modulation of a highly conserved sialometabolism gene cluster in Haemophilus influenzae and its effect on virulence. BMC Microbiol. 10: 48-10.1186/1471-2180-10-48.
Na HS, Kim HJ, Lee HC, Hong Y, Rhee JH, Choy HE: Immune response induced by Salmonella typhimurium defective in ppGpp synthesis. Vaccine. 2006, 24 (12): 2027-2034. 10.1016/j.vaccine.2005.11.031.
Morona R, van den Bosch L, Manning PA: Molecular, genetic, and topological characterization of O-antigen chain length regulation in Shigella flexneri. J Bacteriol. 1995, 177 (4): 1059-1068.
Menard R, Sansonetti PJ, Parsot C: Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J Bacteriol. 1993, 175 (18): 5899-5906.
Miki T, Okada N, Danbara H: Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem. 2004, 279 (33): 34631-34642. 10.1074/jbc.M402760200.
Sternberg NL, Maurer R: Bacteriophage-mediated generalized transduction in Escherichia coli and Salmonella typhimurium. Methods Enzymol. 1991, 204: 18-43. full_text.
Datsenko KA, Wanner BL: One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000, 97 (12): 6640-6645. 10.1073/pnas.120163297.
Kuruma H, Egawa S, Oh-Ishi M, Kodera Y, Satoh M, Chen W, Okusa H, Matsumoto K, Maeda T, Baba S: High molecular mass proteome of androgen-independent prostate cancer. Proteomics. 2005, 5 (4): 1097-1112. 10.1002/pmic.200401115.
Miller JH: A Short Course in Bacterial Genetics. 1992, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 72-74.
Gotoh H, Okada N, Kim YG, Shiraishi K, Hirami N, Haneda T, Kurita A, Kikuchi Y, Danbara H: Extracellular secretion of the virulence plasmid-encoded ADP-ribosyltransferase SpvB in Salmonella. Microb Pathog. 2003, 34 (5): 227-238. 10.1016/S0882-4010(03)00034-2.
Simon R, Priefer U, Puhler A: A Braod host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983, 1: 784-791. 10.1038/nbt1183-784.
Acknowledgements
We thank Toru Hattori (SCRUM inc, Japan) for 2-DE gel image analysis. We thank Kaori Dobashi, Nobue Nameki, Masato Hosono, Kohei Yamashita, and Ayako Mizuta for their technical assistance.
This work was supported in part by Grants-in-Aid for Young Scientists (B) (17790291 and 22790415 for TH) and for Scientific Research (C) (17590398 and 21590490 for NO) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, and by a Kitasato University Research Grant for Young Researchers (2010 for TH).
Author information
Authors and Affiliations
Corresponding author
Additional information
Authors' contributions
TH, SM, YYO, YKo, and SSI performed the experiments. TH and NO designed the experiments. TMi constructed the TM157, TM129, and TM548 strains. YKu assisted with the experiments. MOI, TMa, and HD advised regarding the design of the experiments. TH and NO wrote the paper.
Electronic supplementary material
Authors’ original submitted files for images
Below are the links to the authors’ original submitted files for images.
Rights and permissions
Open Access This article is published under license to BioMed Central Ltd. This is an Open Access article is distributed under the terms of the Creative Commons Attribution License ( https://creativecommons.org/licenses/by/2.0 ), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Haneda, T., Sugimoto, M., Yoshida-Ohta, Y. et al. Comparative proteomic analysis of Salmonella enterica serovar Typhimurium ppGpp-deficient mutant to identify a novel virulence protein required for intracellular survival in macrophages. BMC Microbiol 10, 324 (2010). https://doi.org/10.1186/1471-2180-10-324
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/1471-2180-10-324