Abstract
Background
Glaucoma is the second leading cause of blindness in the world with primary open-angle glaucoma (POAG) that is the most prevalent type. Brain-derived neurotrophic factor (BDNF) is a member of the neurotrophin family synthesized by retinal ganglion cells (RGCs). Disturbance of axonal transport of neurotrophins with optic nerve dystrophy results in deprivation of BDNF support to the RGCs inducing glaucomatous retinal cell death.
Materials and methods
This case-control study was conducted on 50 POAG patients (mean age 55 ± 10) and 50 healthy control subjects (mean age 40 ± 11). Both groups underwent full ophthalmological examination. Genomic DNA was extracted followed by BDNF rs2030324 genotyping by real time PCR.
Results
Correlation coefficient analysis showed significant positive correlation between age and right and left cup to disc ratio (r = 0.448, p = 0.001; r = 0.283, p = 0.004 respectively) and significant negative correlation between intraocular pressure and right and left VA (r = − 0.212, p = 0.034; r = − 0.258, p = 0.009 respectively). No significant difference between the 2 groups was found as regards genotype or alleles frequency distribution (p = 0.722).
Conclusion
This study did not succeed to illustrate the role of BDNF gene polymorphism (SNP rs2030324) as a risk factor for POAG occurrence. The mechanism of glaucoma development according to the BDNF polymorphism remains unclear.
Similar content being viewed by others
Introduction
Glaucoma is a chronic neurodegenerative disorder characterized by damage of retinal ganglion cells (RGCs) and their axons with deformation of the optic nerve head resulting in an irreversible loss of vision (Liu et al. 2018). Glaucoma is the second leading cause of blindness in the world. There are about 60 million people worldwide with glaucomatous optic neuropathy of which 8.4 million are blind. The global incidence of glaucoma is anticipated to increase to 76 million by 2020 and 111.8 million by 2040 (Tham et al. 2014). All forms of the disease have in common a characteristic potentially progressive optic neuropathy that is associated with visual field loss as damage progresses, and in which intraocular pressure (IOP) is a key modifiable factor (Brad Bowling 2016).
Death of RGCs is not completely understood, and the sole target for clinical intervention is increased IOP or ocular hypertension (Vidal-Sanz et al. 2012). Unfortunately, despite the reduction of IOP by medical or surgical treatment, progressive loss of vision is still noticeable. For this reason, there must be an IOP-independent mechanism, encouraging researchers to search for alternative treatment strategies aiming for neuroprotection of the retinal ganglion cell (Gupta and Yucel 2007).
The underlying cause of RGCs death in neurodegenerative diseases seems to be apoptosis; a programmed cell death. Processes that lead to apoptosis in the development of glaucoma include glutamate excite toxicity, blockage of axonal transport, antibodies to heat shock proteins, and ischemia (Nowak et al. 2015).
Normal axonal transport is vital to the well functioning of neurons, while retrograde transport of neurotrophins may be necessary for the survival of RGCs. One of the neurotrophins which shows a role in retrograde axonal transport is brain-derived neurotrophic factor (BDNF), which is important to the growth and survival of nerve cells (Pease, ME et al 2000; Kuehn et al. 2005).
In animal models of glaucoma, BDNF delivery to the retina is substantially reduced. Meanwhile, Pease and co-workers in 2000 have shown that the injection of BDNF into the vitreous cavity of rats with experimentally elevated IOP leads to increased cell survival of RGCs, compared with untreated eyes.
The BDNF gene is a protein-coding gene located on chromosome11p14.1; it contains 11 exons, 9 of which contain a specific promoter that regulates its expression (Modarresi et al. 2012). The rs2030324 SNP is in a non-coding intron region of the BDNF gene, it is T-to-C substitution. The biological mechanisms of the rs2030324 effects are not fully known but could potentially involve some effects on BDNF function in neurons as genetic associations involving rs2030324in haplotypes have been reported, and also it was found to be associated with multiple sclerosis (Guttman Weinstock et al. 2011).
This study was conducted to assess the role of the brain-derived neurotrophic factor (BDNF) gene polymorphism (rs2030324) in primary open-angle glaucoma (POAG) patients.
Patients and methods
Selection of patients
This case-control study was performed in the Chemical Pathology Department, Faculty of Medicine, Cairo University in collaboration with the Research Institute of Ophthalmology Hospital, from January 2016 until June 2017 performed on 100 subjects, non-familial related, recruited from outpatient clinic at the Research Institute of Ophthalmology.
The subjects were classified into two groups: group 1 included 50 patients with primary open-angle glaucoma (POAG) showing optic nerve and visual field changes compatible with glaucomatous damage; group 2 included 50 healthy control subjects without glaucoma symptoms with normal IOP, no changes in the optic nerve after examination of the fundus and no changes in the structure of the anterior and posterior segments of the eye by slit-lamp examination (Table 1). Informed consents were obtained before enrolment into the study.
Exclusion criteria among cases included (a) uveitis, trauma, or secondary glaucoma; (b) use of any eye drops other than anti-glaucoma preparations, present or past treatment with glucocorticoids or immunosuppressive therapy; (c) any ocular surgeries or laser treatments; and (d) diabetics.
Sample collection and storage
Two milliliters were collected in a sterile EDTA vacutainer for DNA extraction. Samples were kept frozen at − 80 °C until the time of analysis.
Methods
All patients and controls were subjected to the following:
-
a)
Complete ophthalmological evaluations that included full medical history, best corrected visual acuity, slit-lamp examination, evaluation of the anterior chamber angle by goniolens, evaluation of IOP using applanation tonometer, fundus examination using the 90 diopter, and automated visual field analysis using 24-2 threshold test (Humphrey).
-
b)
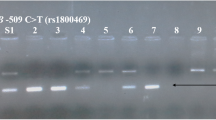
Laboratory investigations: analysis of BDNF genotype rs2030324 by real-time PCR using TaqMan® Probe, ID assay no. 4351379, (Applied Biosystems, Foster City, California, USA), performed on ®Step One Real-time PCR system (Applied Biosystems). It was done in two steps:
Step 1: Extraction of genomic DNA from peripheral blood leucocytes was performed by QIAamp DNA blood mini-kit (Qiagen, Hilden, Germany) which contains silica-gel spin columns (Boom et al. 1990). The concentration of DNA was determined by measuring the absorbance at 260 nm (A260) using the Nanodrop spectrophotometer. The ratio of the readings at 260 nm and 280 nm (A260/A280) provides an estimate of the purity of DNA. DNA used in this study has an A260/A280 ratio of 1.7:1.9.
Step 2: BDNF rs2030324 genotyping by real-time PCR allelic discrimination, TaqMan MGB (minor groove binder) probes assay. The total reaction volume was 20 μl/well using the following amplification protocol: an initial activation of AmpliTaq Gold DNA polymerase for 10 min at 95 °C, followed by 40 PCR cycles, each cycle consisted of: denaturation at 92 °C for 15 s, annealing at 60 °C for 60 s and finally extension at 60 °C for 60 s. The Sequence Detection System (SDS) software used the fluorescence measurements made during the plate read to plot fluorescence (Rn) values based on the signals from each well. The plotted fluorescence signals indicated which alleles were in each sample.
Statistical analysis
Data were statistically described in terms of mean ± standard deviation (± SD), median and (25th–75th percentile), or frequencies and percentage when appropriate. Comparison of numerical variables between the study groups was done using the Student t test for parametric data and Mann–Whitney for nonparametric data in comparing 2 groups. For comparing categorical data, chi-square (χ2) test was performed. Exact test was used instead when the expected frequency is less than 5. P values less than 0.05 was considered statistically significant. Correlation was done using Spearman’s correlation statistics. All statistical calculations were done using computer program SPSS (SPSS Inc., Chicago, IL, USA) release 15 for Microsoft Windows (2006).
Results
The two groups showed significant difference in ocular parameters: right and left VA, IOP, and C/D ratio (P < 0.05) (Table 1), the two groups were sex matched (P = 0.687). BDNF rs2030324 genotypes and alleles distribution between the two groups showed no significant difference (P = 0.722). There was no statistical significance association between BDNF genotypes distribution and any of ophthalmological clinical parameters (P > 0.05) (Table 2).
Spearman’s correlation coefficient analysis showed a statistical significant correlation between the following parameters (Table 3):
-
Age with Rt and Lt visual acuity: significant negative correlation (r = − 0.473, P = 0.001; r = − 0.453, P = 0.001 respectively)
-
Age with Rt and Lt C/D: significant positive correlation (r = 0.448, P = 0.001; r = 0.283 P = 0.004 respectively)
-
Lt IOP with Rt and Lt VA: significant negative correlation (r = − 0.212, P = 0.034; r = − 0.258, P = 0.009 respectively)
-
Rt MD with Lt MD: significant positive correlation (r = 0.729, P = 0.001)
-
Lt C/D with Rt VA: significant negative correlation (r = − 0.253, P = 0.011)
-
Rt C/D with Rt and Lt VA: significant negative correlation (r = − 0.404, P = 0.001; r = − 0.267, P = 0.007 respectively)
-
Rt C/D and Lt C/D: significant positive correlation (r = 0.717, P = 0.001)
-
Lt IOP and Rt IOP: significant positive correlation (r = 0.621 P = 0.001) (Table 1)
Data presented as number and percent (%)
Discussion
Glaucoma along with Alzheimer’s disease (AD), Parkinson’s disease (PD), and multiple sclerosis (MS) is classified as a neurodegenerative disorder. There are resemblances in cellular events that lead to the development of glaucoma in the aforementioned diseases. Moreover, optic nerves from AD patients are characterized by the loss of RGCs, the earliest dying cells in glaucoma (Nowak et al. 2015). POAG accounts for three-quarters (74%) of all glaucoma cases and has a complex hereditary component; it has been associated with at least 20 loci in the human genome (Brad Bowling 2016).
BDNF is one of the key neurotrophic factors in glaucoma that is reduced in the optic nerve head and serum of POAG patients, suggesting that BDNF may be a biomarker for glaucoma. It is encoded by BDNF gene on chromosome11p14.1 (Kimura et al. 2016). BDNF mediates protection from apoptosis by p53 activation and exerts its pro-survival effects by binding to its receptor TrkB (Tropomyosin receptor kinase B), activating signaling pathways (Gupta et al. 2014). In animal models of glaucoma, BDNF delivery to the retina is reduced. However, injection of BDNF into the vitreous cavity of rats with experimentally elevated IOP leads to increased cell survival of RGCs (Nowak et al. 2015).
The rs2030324 polymorphism is located in the promoter site of BDNF gene; hence, it might have an influence on gene expression. Up to our knowledge, the rs2030324 SNP has not been investigated in patients of POAG in Egypt, neither on a wide nor on a narrow scale.
In the current study, Rt and Lt IOP and Rt and Lt C/D were significantly higher among glaucomatous patients than that among controls (P = 0.004 and P = 0.004 respectively). However, there was no statistical significant association between clinical parameters of both Rt and Lt (VA, IOP, C/D, and MD) and different BDNF genotypes SNP rs2030324 (P > 0.05). Moreover, there was no significance difference in BDNF genotypes or alleles frequencies between glaucoma patients and controls.
In contrast to our results, Nowak et al. (2015) found a significant higher frequency of the TT, TC genotype, and T allele of BDNF SNP rs2030324 (P < 0.001) in patients with POAG compared with the controls. Their study was performed by PCR-RFLP method on a large cohort of 769 unrelated Caucasian polish subjects: 363 POAG patients and 406 healthy controls. Their results showed association between BDNF genotypes and disease progression, where they found an increase in C/D in associated with TT BDNF genotype (P = 0.004).
This discrepancy between results may be due to different ethnic groups, racial differences, sample size, poorly characterized controls, and use of different methods for genotyping.
By analysis of BDNF expression level in the same study, Nowak et al. (2015) found no significant differences in serum BDNF mRNA expression levels between POAG patients and the controls (P > 0.05), while analysis of the BDNF expression level in relation to the clinical parameters showed a decrease of mRNA expression levels with a decrease of the RA value (rim area) (P = 0.011). RA reflects the real amount of nerve fibers in optic disc, which may indicate the protective role of BDNF in the development of glaucoma.
Another polymorphic site (196G/A) of BDNF gene was evaluated by Nowak et al. (2014) on 362 unrelated Caucasian polish subjects (169 POAG patients and 193 healthy controls). They found no significant association in frequency of BDNF (196G/A) genotypes or alleles and POAG. It seems that other polymorphic variants of BDNF gene may be involved in the development of POAG.
Conclusion
To the contrary of numerous data suggesting a role of BDNF in the pathogenesis and progression of glaucoma, this study did not succeed to illustrate the role of BDNF gene polymorphism (SNP rs2030324) as a risk factor for POAG occurrence. The mechanism of glaucoma development according to the BDNF polymorphism remains unclear, so we recommend (1) further studying of BDNF gene polymorphism SNP rs2030324 on larger sample size, (2) studying other SNPs of BDNF gene that may have a role in POAG, and (3) using DNA sequencing techniques for detection of different BDNF gene polymorphisms and other genes involved in the neurodegeneration mechanism for better understanding of the molecular basis of POAG and to find early diagnostic markers of POAG development
Availability of data and materials
All authors declare that the data and materials are available.
Abbreviations
- BDNF:
-
Brain-derived neurotrophic factor
- POAG:
-
Primary open-angle glaucoma
- RGCs:
-
Retinal ganglion cells
- VA:
-
Visual acuity
References
Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J (1990) Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28(3):495–503
Brad Bowling (2016): Kanski’s textbook, a systematic approach, 8th edition, Elsevier Limited, chapter 10 Glaucoma, p305:358.
Gupta N, Yucel YH (2007) Glaucoma as a neurodegenerative disease. Curr Opin Ophthalmol 18(2):110–114
Gupta V, You Y, Li J, Gupta V, Golzan M, Klistorner A, van den Buuse M, Graham S (2014) BDNF impairment is associated with age-related changes in the inner retina and exacerbates experimental glaucoma. Biochim.Biophys.Acta 1842:1567–1578
Guttman Weinstock B, Benedict RH, Tamano-Blanco M, Ramasamy DP, Stosic M, Polito J, Zivadinov R, Ramanathan M (2011) The rs2030324 SNP of brain-derived neurotrophic factor (BDNF) is associated with visual cognitive processing in multiple sclerosis. Pathophysiology. 18(1):43–52
Kimura A, Kazuhiko N, Guo X, Chikako H, Takayuki H (2016) Neuroprotection, growth factors and BDNF-TrkB signaling in retinal degeneration. Int J Mol Sci 17(9):1584
Kuehn MH, Fingert JH, Kwon YH (2005) Retinal ganglion cell death in glaucoma: mechanisms and neuroprotective strategies. Ophthalmol Clin N Am 18(3):383–395
Liu B, McNally S , Kilpatrick J I, Jarvis S P , O'Brien C J. (2018):Aging and ocular tissue stiffness in glaucoma .January February, 2018Volume 63, Issue 1, Pages 56–74.
Modarresi F, Faghihi MA, Lopez-Toledano MA, Fatemi RP, Magistri M, Brothers SP et al (2012) Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional up regulation. Nat Biotechnol 30(5):453–459
Nowak A , Majsterek I , Przybyłowska-Sygut K , Pytel D , Szymanek K , Szaflik J & Szaflik JP (2015): Analysis of the expression and polymorphism of APOE, HSP, BDNF, and GRIN2B genes associated with the neurodegeneration process in the pathogenesis of primary open angle glaucoma. BioMed Research International, 258281.
Nowak A, Szaflik JP, Gacek M, Przybylowska-Sygut K, Kamińska A, Szaflik J, Majsterek I (2014) BDNF and HSP gene polymorphisms and their influence on the progression of primary open-angle glaucoma in a Polish population. Archives of Medical Science: AMS 10(6):1206–1213
Pease ME, McKinnon SJ, Quigley HA, Kerrigan-Baumrind LA, Zack DJ (2000) Obstructed axonal transport of BDNF and its receptor TrkB in experimental glaucoma. Invest Ophthalmol Vis Sci 4:764–774
Tham Y-CC, Li X, Wong TY, Quigley HA, Aung T, Ed F, Cheng CY (2014) Global prevalence of glaucoma and projections of glaucoma burden through 2040. A systematic review and meta-analysis. Ophthalmology 121:2081–2090
Vidal-Sanz M, Salinas-Navarro M, Nadal-Nicolas FM, Alarcon-Martinez L, Valiente-Soriano FJ, de Imperial JM et al (2012) Understanding glaucomatous damage anatomical and functional data from ocular hypertensive rodent retinas. ProgRetin Eye Res 31:1–27
Disclosure statement
The authors declare that they do not have conflict of interest.
Equipment | Source |
1-Pcr (TProfessional Basic) | Research Institute of Ophthmology (RIO) |
2-Gel electrophoresis unit used in this study was provided by Biometra, Germany (Model: Compact M). | Research Institute of Ophthmology (RIO) |
Kit | Source |
1-Blood sample | Research Institute of Ophthmology (RIO) |
2-DNA extraction | (Qiagen, Hilden, Germany |
3-Dream Taq DNA polymerase | Thermo Scientific |
4-primers | Thermo Scientific |
5-Agarose A and Tris-Borate-EDTA (TBE) | Biobasic, Canada |
6-GeneRuler 20 bp | Thermo Scientific |
Funding
The authors declare that we have not any fund.
Author information
Authors and Affiliations
Contributions
MF developed the research idea of scientific and legal aspects and supervising the practical part carried studies out the molecular genetics. AMD supervised the part of the biochemistry practical and theoretical work, participated in the sequence alignment and drafted the manuscript. SS supervised the part of the practical and theoretical genetic work, carried out the immunoassays and revised the manuscript. HMEH worked the practical and theoretical part of the research, participated in the sequence alignment and drafted the manuscript. FMG worked the practical and theoretical part of the research, participated in the design of the study. IAF diagnosed patients of diabetic retinopathy, performed the statistical analysis, and helped to revise the manuscript. SMH diagnosed patients of diabetic retinopathy conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The study was approved by the ethical committee of the Research Institute of Ophthalmology.
Consent for publication
All authors declare that they approved the publication of this research in the Bulletin of National Research Center.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fathy, M., Darweesh, A.M., Sharaf, S. et al. Brain-derived neurotrophic factor (BDNF) gene polymorphism in a cohort of Egyptian primary open-angle glaucoma (POAG) patients. Bull Natl Res Cent 44, 45 (2020). https://doi.org/10.1186/s42269-020-00288-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s42269-020-00288-x