Abstract
Background
Macrosomia is defined as an infant’s birth weight of more than 4000 g. Although microRNAs (miRNAs) have been implicated in the pathogenesis of various diseases, the associations between serum miRNAs and macrosomia have been rarely reported.
Methodology
We used the Taqman Low Density Array followed by quantitative reverse transcriptase polymerase chain reaction assays to screen for miRNAs associated with macrosomia using serum samples collected 1 week before delivery.
Results
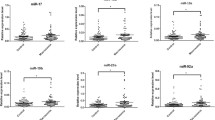
Profiling results showed that 1 miRNA was significantly upregulated and 10 miRNAs were significantly downregulated in serum samples of macrosomia (ΔΔCt > 3-fold). The expression levels of miR-21 were significantly decreased in macrosomia as compared to the controls in the third trimester. Receiver operating characteristic (ROC) curve analyses showed that the area under the ROC curve for miR-21 was 67.7% (sensitivity = 66.7% and specificity = 70.0%).
Conclusions
miR-21 in maternal serum is differentially expressed between macrosomia and controls, and miR-21 could be used as a candidate biomarker to predict macrosomia.
Similar content being viewed by others
References
Lu Y, Zhang J, Lu X, Xi W, Li Z. Secular trends of macrosomia in southeast China, 1994–2005. BMC Public Health. 2011;11:818.
Koyanagi A, Zhang J, Dagvadorj A, et al. Macrosomia in 23 developing countries: an analysis of a multicountry, facility-based, cross-sectional survey. Lancet. 2013;381(9865):476–483.
Xie RH, Cao H, Hong B, Sprague AE, Walker M, Wen SW. Occurrence and predictors of vacuum and forceps used sequentially for vaginal birth. J Obstet Gynaecol Can. 2013;35(4): 317–322.
Yu ZB, Han SP, Zhu GZ, Zhu C, Wang XJ, et al. Birth weight and subsequent risk of obesity: a systematic review and meta-analysis. Obes Rev. 2011;12(7):525–542.
Li XY, Jiang Y, Hu N, et al. [Prevalence and characteristic of overweight and obesity among adults in China, 2010]. Zhonghua Yu Fang YiXue Za Zhi. 2012;46(8):683–686.
Cui Z, Huxley R, Wu Y, Dibley MJ. Temporal trends in overweight and obesity of children and adolescents from nine Provinces in China from 1991–2006. Int J Pediatr Obes. 2010;5(5): 365–374.
Gu S, An X, Fang L, et al. Risk factors and long-term health consequences of macrosomia: a prospective study in Jiangsu Province, China. J Biomed Res. 2012;26(4):235–240.
Yu Z, Han S, Zhu J, Sun X, Ji C, Guo X. Pre-pregnancy body mass index in relation to infant birth weight and offspring overweight/ obesity: a systematic review and meta-analysis. PLoS One. 2013; 8(4):e61627.
Li S, Rosenberg L, Palmer JR, Phillips GS, Heffner LJ, Wise LA. Central adiposity and other anthropometric factors in relation to risk of macrosomia in an African American population. Obesity (Silver Spring). 2013;21(1):178–184.
Grundt JH, Nakling J, Eide GE, Markestad T. Possible relation between maternal consumption of added sugar and sugar-sweetened beverages and birth weight–time trends in a population. BMC Public Health. 2012;12:901.
Silveira PP, Portella AK, Goldani MZ, Barbieri MA. Developmental origins of health and disease (DOHaD). J Pediatr (Rio J). 2007; 83(6):494–504.
Lillycrop KA. Effect of maternal diet on the epigenome: implications for human metabolic disease. Proc Nutr Soc. 2011;70(1):64–72.
Chim SS, Shing TK, Hung EC, et al. Detection and characterization of placental microRNAs in maternal plasma. Clin Chem. 2008;54(3):482–490.
Zhao C, Dong J, Jiang T, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6:e23925.
Enquobahrie DA, Abetew DF, Sorensen TK, et al. Placental microRNA expression in pregnancies complicated by preeclampsia. Am J Obstet Gynecol. 2011;204(2):178. e112–e121.
Jiang H, Wu W, Zhang M, et al. Aberrant upregulation of miR-21 in placental tissues of macrosomia. J Perinatol. 2014;34(9):658–663.
Lawrie CH, Gal S, Dunlop HM, et al. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008; 141(5):672–675.
Hu Z, Chen X, Zhao Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–1726.
Schrauder MG, Strick R, Schulz-Wendtland R, et al. Circulating micro-RNAs as potential blood-based markers for early stage breast cancer detection. PLoS One. 2012;7(1):e29770.
Zhu C, Ren C, Han J, et al. A five-micro RNA panel in plasma was identified as potential biomarker for early detection of gastric cancer. Br J Cancer. 2014;110(9):2291–2299.
Catalano PM. Management of obesity in pregnancy. Obstet Gynecol. 2007;109(2 pt 1):419–433.
Canavan TP, Hill LM. Sonographic biometry in the early third trimester: a comparison of parameters to predict macrosomia at birth [published online September 5, 2014]. J Clin Ultrasound. 2014. doi: 10.1002/jcu.22230.
Su L, Zhao S, Zhu M, Yu M. Differential expression of microRNAs in porcine placentas on days 30 and 90 of gestation. Reprod Fertil Dev. 2010;22(8):1175–1182.
Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518.
Xie Y, Todd NW, Liu Z, et al. Altered miRNA expression in sputum for diagnosis of non-small cell lung cancer. Lung Cancer. 2010;67(2):170–176.
Chen X, Ba Y, Ma L, et al. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18(10):997–1006.
Shen J, Todd NW, Zhang H, et al. Plasma microRNAs as potential biomarkers for non-small-cell lung cancer. Lab Invest. 2011; 91(4):579–587.
Barad O, Meiri E, Avniel A, et al. MicroRNA expression detected by oligonucleotide microarrays: system establishment and expression profiling in human tissues. Genome Res. 2004;14(12): 2486–2494.
Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773.
Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13(1):39–53.
Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10(12):1957–1966.
Volinia S, Calin GA, Liu CG, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103(7):2257–2261.
Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107(6):810–817.
Kim YJ, Hwang SH, Cho HH, Shin KK, Bae YC, Jung JS. MicroRNA 21 regulates the proliferation of human adipose tissue-derived mesenchymal stem cells and high-fat diet-induced obesity alters microRNA 21 expression in white adipose tissues. J Cell Physiol. 2012;227(1):183–193.
Maccani MA, Padbury JF, Marsit CJ. miR-16 and miR-21 expression in the placenta is associated with fetal growth. PLoS One. 2011;6(6):e21210.
Mouillet JF, Chu T, Hubel CA, Nelson DM, Parks WT, Sadovsky Y. The levels of hypoxia-regulated microRNAs in plasma of pregnant women with fetal growth restriction. Placenta. 2010;31(9): 781–784.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Jiang, H., Wen, Y., Hu, L. et al. Serum MicroRNAs as Diagnostic Biomarkers for Macrosomia. Reprod. Sci. 22, 664–671 (2015). https://doi.org/10.1177/1933719114561557
Published:
Issue Date:
DOI: https://doi.org/10.1177/1933719114561557