Abstract
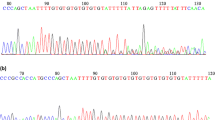
The expression of DNA mismatch repair (DMMR) genes in patients with maturation arrest (MA) was analyzed. Samples were subjected to mutL homolog 3 (MLH3) mutation analysis by denaturing high-performance liquid chromatography/sequencing and quantification of MMR expression in testicular tissue by real-time polymerase chain reaction (PCR). Microsatellite instability assays were negative. Two missense and 1 intronic mutations were found. The missense mutation 2531C/T (P844 L), predicted to affect MLH3 function, was found in 3 MA cases in association with the intronic variant IVS9 + 66G/A. Relative messenger RNA (mRNA) quantification identified 2 patients who overexpressed MLH3, 1 of them also overexpressing mutL homolog 1 (MLH1). The latter also presented the 2531C/T-IVS9 + 66G/A mutation. In conclusion, we suggest that a predominance of MLH3 expression might favor the MLH1/MLH3 complex which then would compete with the MLH1/PMS2 complexes. This could convey disruption of the relative stoichiometry between MLH1/MLH3 and MLH1/PMS2 complexes, thus causing meiosis failure, as MLH1/PMS2 complexes are supposed to replace MLH1/MLH3 during diplonema.
Similar content being viewed by others
References
Vogt PH, Edelmann A, Kirsch S, et al. Human Y chromosome azoospermia factors (AZF) mapped to different subregions in Yq11. Hum Mol Genet. 1996;5(7):933–943.
Sá R, Neves R, Fernandes S, et al. Cytological and expression studies and quantitative analysis of the temporal and stage specific effects of follicle stimulating hormone and testosterone during cocultures of the normal human seminiferous epithelium. Biol Reprod. 2008;79(5):962–975.
Sousa M, Cremades N, Silva J, et al. Predictive value of testicular histology in secretory azoospermic subgroups and clinical outcome after microinjection of fresh and frozen-thawed sperm and spermatids. Hum Reprod. 2002;17(7):1800–1810.
Sá R, Sousa M, Cremades N, Silva J, Barros A. Spermatid injection: current status. In: Rizk B, Garcia-Velasco JA, Sallam HN, Makrigiannakis A, eds. Textbook of Infertility and Assisted Reproduction (MSRM: Mediterranean Society for Reproductive Medicine). London: Cambridge University Press; Chapter 53, 2008:491–503.
Habermann B, Mi H-F, Edelmann A, et al. DAZ (Deleted in AZoospermia) genes encode proteins located in human late spermatids and in sperm tails. Hum Reprod. 1998;13(2):363–369.
Fernandes S, Huellen K, Gonc¸alves J, et al. High frequency of DAZ1/DAZ2 gene deletions in patients with severe oligozoospermia. Mol Hum Reprod. 2002;8(3):286–298.
Ferrás C, Fernandes S, Marques CJ, et al. AZF and DAZ gene copy specific deletion analysis in maturation arrest and Sertoli cell only syndrome. Mol Hum Reprod. 2004;10(10):755–761.
Calogero AE, Garofalo MR, Barone N, et al. Spontaneous regression over time of the germinal epithelium in a Y chromosome-microdeleted patient. Hum Reprod. 2001;16(9):1845–1848.
Lipkin SM, Wang V, Stoler DL, et al. Germline and somatic mutation analyses in the DNA mismatch repair gene MLH3: evidence for somatic mutation in colorectal cancers. Hum Mut. 2001; 17(5):389–396.
Kane MF, Loda M, Gaida GM, et al. Methylation of the hMLH1 promoter correlates with lack of expression of in sporadic colon tumors and mismatch repair-defective human tumor cels. Cancer Res. 1997;57(5):808–811.
Aaltonen LA, Salovaara R, Kristo P, et al. Incidence of Hereditary nonpolyposis colorectal cancer and the feasibility of molecular screening for the disease. N Engl J Med. 1998;338(21):1481–1487.
Cannavo E, Marra G, Sabates-Bellver J, et al. Expression of the Mutl homologue hMLH3 in human cells and its role in DNA mismatch repair. Cancer Res. 2005;65(23):10759–10766.
Lipkin SM, Wang V, Jacoby R, et al. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet. 2000;24(1):27–35.
Loukola A, Vikki S, Singh J, Launonen V, Aaltonen LA. Germline and somatic mutation analysis of MLH3 in MSI-positive colorectal cancer. Am J Pathol. 2000;157(2):347–352.
Akiyama Y, Nagasaki H, Nakajima T, Sakai H, Nomizu T, Yuasa Y. Infrequent frameshift mutations in the simple repeat sequences of hMLH3 in hereditary nonpolyposis colorectal cancers. Jpn J Clin Oncol. 2001;31(2):61–64.
Wu Y, Berends MJ, Sijmons RH, et al. A role for MLH3 in hereditary nonpolyposis colorectal cancer. Nat Genet. 2001;29(2): 137–138.
Hienonem T, Laiho P, Salovaara R, et al. Little evidence for involvement of MLH3 in colorectal cancer predisposition. Int J Cancer. 2003;106(2):292–296.
Liu HX, Zhou XL, Liu T, et al. The role of hMLH3 in familial colorectal cancer. Cancer Res, 2003;63(8):1894–1899.
Muramatsu M, Kinoshita K, Fagarasan S, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102(5):553–563.
Revy P, Muto T, Levy Y, et al. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the hyper-IgM syndrome (HIGM2). Cell. 2000;102(5):565–575.
Li Z, Scherer SJ, Ronai D, et al. Examination of Msh6- and Msh3-deficient mice in class switching reveals overlapping and distinct roles of Muts homologues in antibody diversification. J Exp Med. 2004;200(1):47–59.
Li Z, Peled JU, Zhao C, et al. A role for Mlh3 in somatic hyper-mutation. DNA Repair. 2006;5(6):675–682.
Kondo E, Horii A, Fukushige S. The interacting domains of three MutL heterodimers in man: hMLH1 interacts with 36 homologous amino acid residues within hMLH3, hPMS1 and hPMS2. Nucleic Acids Res. 2001;29(8):1695–1702.
Chen PC, Dudley S, Hahen W, et al. Contributions be Mutl homologues Mlh3 and Pms2 to DNA mismatch repair and tumor suppression in the mouse. Cancer Res. 2005;65(19):8662–8670.
Lipkin SM, Moens PB, Wang V, et al. Meiotic arrest and aneuploidy in MLH3-deficient mice. Nat Genet. 2002;31(4): 385–390.
Santucci-Darmanin S, Neyton S, Lespinasse F, Saunieres A, Gaudray P, Paquis-Flucklinger V. The DNA mismatch-repair MLH3 protein interacts with MSH4 in meiotic cells, supporting a role for this MutL homolog in mammalian meiotic recombination. Hum Mol Genet. 2002;11(15):1697–1706.
Marcon E, Moens P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics. 2003;165(4):2283–2287.
Boland CR, Thibodeau SN, Hamilton SR, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257.
Ferrás C, Zhou XL, Sousa M, Lindblom A, Barros A. DNA mismatch repair gene hMLH3 variants in meiotic arrest. Fertil Steril. 2007;88(6):1681–1684.
Sousa M, Cremades N, Alves C, et al. Developmental potential of human spermatogenic cells co-cultered with Sertoli cells. Hum Reprod. 2002;17(1):161–172.
Taylor NP, Powell MA, Gibb RK, et al. MLH3 mutation in endometrial cancer. Cancer Res. 2006;66(15):7502–7508.
Baker SM, Bronner CE, Zhang L, et al. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82(2):309–319.
Edelmann W, Cohen PE, Kane M, et al. Meiotic pachytene arrest in MLH1-deficient mice. Cell. 1996;85(7):1125–1134.
Edelmann W, Cohen PE, Kneitz B, et al. Mammalian MutS homologue 5 is required for chromosome pairing in meiosis. Nat Genet. 1999;21(1):123–127.
Kneitz B, Cohen PE, Avdievich E, et al. MutS homolog 4 localization to meiotic chromosomes is required for chromosome pairing during meiosis in male and female mice. Genes Dev. 2000;14(9):1085–1097.
Nudell D, Castillo M, Turek PJ, Pera RR. Increased frequency of mutations in DNA from infertile men with meiotic arrest. Hum Reprod. 2000;15(6):1289–1294.
Terribas E, Bonache S, Garcia-Arevalo M, et al. Changes in the expression profile of the meiosis-involved mismatch repair genes in impaired human spermatogenesis. J Androl. 2010;31(4):346–357.
Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ferrás, C., Fernandes, S., Silva, J. et al. Expression Analysis of MLH3, MLH1, and MSH4 in Maturation Arrest. Reprod. Sci. 19, 587–596 (2012). https://doi.org/10.1177/1933719111428521
Published:
Issue Date:
DOI: https://doi.org/10.1177/1933719111428521