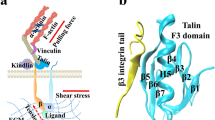
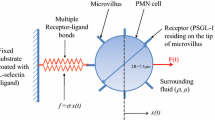
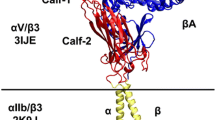
Abstract
Integrin αIIbβ3 expresses on the plasma membrane of platelets, which can be activated by mechanical pulling force from its ligand. Herein, we performed all-atom molecular dynamics (MD) simulations on the full-length integrin embedded in the membrane with a ligand, fibronectin (FN), attaching to the integrin head. A pulling force was applied to the ligand in the MD simulations. The force applied on the ligand could transmit to the integrin head through the transient interaction, and have chances to induce the extension of the integrin. The loading rate affects the pulling force but has limited effects on the integrin extension. Our simulation showed the mechanical force could induce the conformational change of the membrane-embedded integrin.






Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are not publicly available due to the extremely large data volume but are available from the corresponding author upon reasonable request.
References
M. Humphries, Integrin structure. Biochem. Soc. Trans. 28, 311–340 (2000)
M. Shimaoka, J. Takagi, T.A. Springer, Conformational regulation of integrin structure and function. Annu. Rev. Biophys. Biomol. Struct. 31, 485–516 (2002)
J.E. Meredith, M.A. Schwartz, Integrins, adhesion and apoptosis. Trends Cell Biol. 7, 146–150 (1997)
J.-P. Xiong, T. Stehle, B. Diefenbach, R. Zhang, R. Dunker, D.L. Scott, A. Joachimiak, S.L. Goodman, M.A. Arnaout, Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345 (2001)
I.D. Campbell, M.J. Humphries, Integrin structure, activation, and interactions. Cold Spring Harb. Perspect. Biol. 3, a004994 (2011)
J. Li, Y. Su, W. Xia, Y. Qin, M.J. Humphries, D. Vestweber, C. Cabañas, C. Lu, T.A. Springer, Conformational equilibria and intrinsic affinities define integrin activation. EMBO J. 36, 629–645 (2017)
T. Schürpf, T.A. Springer, Regulation of integrin affinity on cell surfaces. EMBO J. 30, 4712–4727 (2011)
J. Takagi, B.M. Petre, T. Walz, T.A. Springer, Global conformational rearrangements in integrin extracellular domains in outside-in and inside-out signaling. Cell 110, 599–611 (2002)
T. Xiao, J. Takagi, B.S. Coller, J.-H. Wang, T.A. Springer, Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67 (2004)
Z. Sun, S.S. Guo, R. Fässler, Integrin-mediated mechanotransduction. J. Cell Biol. 215, 445–456 (2016)
S.J. Holland, N.W. Gale, G. Mbamalu, G.D. Yancopoulos, M. Henkemeyer, T. Pawson, Bidirectional signalling through the EPH-family receptor Nuk and its transmembrane ligands. Nature 383, 722–725 (1996)
R.O. Hynes, Integrins: bidirectional, allosteric signaling machines. Cell 110, 673–687 (2002)
R. Alon, M.L. Dustin, Force as a facilitator of integrin conformational changes during leukocyte arrest on blood vessels and antigen-presenting cells. Immunity 26, 17–27 (2007)
E. Puklin-Faucher, M.P. Sheetz, The mechanical integrin cycle. J. Cell Sci. 122, 179–186 (2009)
W. Chen, J. Lou, E.A. Evans, C. Zhu, Observing force-regulated conformational changes and ligand dissociation from a single integrin on cells. J. Cell Biol. 199, 497–512 (2012)
R. Kolasangiani, T.C. Bidone, M.A. Schwartz, Integrin conformational dynamics and mechanotransduction. Cells 11, 3584 (2022)
F. Kong, A.J. García, A.P. Mould, M.J. Humphries, C. Zhu, Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009)
Y. Chen, H. Lee, H. Tong, M. Schwartz, C. Zhu, Force regulated conformational change of integrin αVβ3. Matrix Biol. 60, 70–85 (2017)
A. Elosegui-Artola, R. Oria, Y. Chen, A. Kosmalska, C. Pérez-González, N. Castro, C. Zhu, X. Trepat, P. Roca-Cusachs, Mechanical regulation of a molecular clutch defines force transmission and transduction in response to matrix rigidity. Nat. Cell Biol. 18, 540–548 (2016)
W. Chen, J. Lou, C. Zhu, Forcing switch from short-to intermediate-and long-lived states of the αA domain generates LFA-1/ICAM-1 catch bonds. J. Biol. Chem. 285, 35967–35978 (2010)
Y.I. Choi, J.S. Duke-Cohan, W. Chen, B. Liu, J. Rossy, T. Tabarin, L. Ju, J. Gui, K. Gaus, C. Zhu, Dynamic control of β1 integrin adhesion by the plexinD1-sema3E axis. Proc. Natl. Acad. Sci. 111, 379–384 (2014)
F. Rosetti, Y. Chen, M. Sen, E. Thayer, V. Azcutia, J.M. Herter, F.W. Luscinskas, X. Cullere, C. Zhu, T.N. Mayadas, A lupus-associated Mac-1 variant has defects in integrin allostery and interaction with ligands under force. Cell Rep. 10, 1655–1664 (2015)
D. Li, B. Ji, Predicted rupture force of a single molecular bond becomes rate independent at ultralow loading rates. Phys. Rev. Lett. 112, 078302 (2014)
N. Plattner, S. Doerr, G. De Fabritiis, F. Noé, Complete protein–protein association kinetics in atomic detail revealed by molecular dynamics simulations and Markov modelling. Nat. Chem. 9, 1005–1011 (2017)
C. Rakers, M. Bermudez, B.G. Keller, J. Mortier, G. Wolber, Computational close up on protein–protein interactions: how to unravel the invisible using molecular dynamics simulations? Wiley Interdiscip. Rev. Comput. Mol. Sci. 5, 345–359 (2015)
X. Li, Z. Fang, D. Li, Z. Li, Binding kinetics study of SARS-CoV-2 main protease and potential inhibitors via molecular dynamics simulations. Phys. Chem. Chem. Phys. 25, 15135–15145 (2023)
D. Li, B. Ji, K.-C. Hwang, Y. Huang, Strength of hydrogen bond network takes crucial roles in the dissociation process of inhibitors from the HIV-1 protease binding pocket. PLoS ONE 6, e19268 (2011)
D.E. Shaw, P. Maragakis, K. Lindorff-Larsen, S. Piana, R.O. Dror, M.P. Eastwood, J.A. Bank, J.M. Jumper, J.K. Salmon, Y. Shan, Atomic-level characterization of the structural dynamics of proteins. Science 330, 341–346 (2010)
S. Piana, K. Lindorff-Larsen, D.E. Shaw, Protein folding kinetics and thermodynamics from atomistic simulation. Proc. Natl. Acad. Sci. 109, 17845–17850 (2012)
J.D. Durrant, J.A. McCammon, Molecular dynamics simulations and drug discovery. BMC Biol. 9, 1–9 (2011)
P.-C. Do, E.H. Lee, L. Le, Steered molecular dynamics simulation in rational drug design. J. Chem. Inf. Model. 58, 1473–1482 (2018)
D. Li, M.S. Liu, B. Ji, Mapping the dynamics landscape of conformational transitions in enzyme: the adenylate kinase case. Biophys. J. 109, 647–660 (2015)
D. Li, B. Ji, K. Hwang, Y. Huang, Crucial roles of the subnanosecond local dynamics of the flap tips in the global conformational changes of HIV-1 protease. J. Phys. Chem. B 114, 3060–3069 (2010)
W. Chen, J. Lou, J. Hsin, K. Schulten, S.C. Harvey, C. Zhu, Molecular dynamics simulations of forced unbending of integrin αVβ3. PLoS Comput. Biol. 7, e1001086 (2011)
M. Nagae, S. Re, E. Mihara, T. Nogi, Y. Sugita, J. Takagi, Crystal structure of α5β1 integrin ectodomain: atomic details of the fibronectin receptor. J. Cell Biol. 197, 131–140 (2012)
E. Puklin-Faucher, V. Vogel, Integrin activation dynamics between the RGD-binding site and the headpiece hinge. J. Biol. Chem. 284, 36557–36568 (2009)
L. Levin, E. Zelzion, E. Nachliel, M. Gutman, Y. Tsfadia, Y. Einav, A single disulfide bond disruption in the β3 integrin subunit promotes thiol/disulfide exchange, a molecular dynamics study. PLoS ONE 8, e59175 (2013)
S. Su, Y. Ling, Y. Fang, J. Wu, Force-enhanced biophysical connectivity of platelet β3 integrin signaling through Talin is predicted by steered molecular dynamics simulations. Sci. Rep. 12, 4605 (2022)
T.C. Bidone, A. Polley, J. Jin, T. Driscoll, D.V. Iwamoto, D.A. Calderwood, M.A. Schwartz, G.A. Voth, Coarse-grained simulation of full-length integrin activation. Biophys. J. 116, 1000–1010 (2019)
J. Yang, Y.-Q. Ma, R.C. Page, S. Misra, E.F. Plow, J. Qin, Structure of an integrin αIIbβ3 transmembrane-cytoplasmic heterocomplex provides insight into integrin activation. Proc. Natl. Acad. Sci. 106, 17729–17734 (2009)
S. Jo, T. Kim, W. Im, Automated builder and database of protein/membrane complexes for molecular dynamics simulations. PLoS ONE 2, e880 (2007)
E.L. Wu, X. Cheng, S. Jo, H. Rui, K.C. Song, E.M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R.M. Venable, CHARMM-GUI Membrane Builder Toward Realistic Biological Membrane Simulations (Wiley, New York, 2014)
J. Zhu, B.-H. Luo, T. Xiao, C. Zhang, N. Nishida, T.A. Springer, Structure of a complete integrin ectodomain in a physiologic resting state and activation and deactivation by applied forces. Mol. Cell 32, 849–861 (2008)
J.F. Van Agthoven, J.-P. Xiong, J.L. Alonso, X. Rui, B.D. Adair, S.L. Goodman, M.A. Arnaout, Structural basis for pure antagonism of integrin αVβ3 by a high-affinity form of fibronectin. Nat. Struct. Mol. Biol. 21, 383–388 (2014)
P. Mark, L. Nilsson, Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960 (2001)
M.J. Abraham, T. Murtola, R. Schulz, S. Páll, J.C. Smith, B. Hess, E. Lindahl, GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015)
J. Huang, A.D. MacKerell Jr., CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J. Comput. Chem. 34, 2135–2145 (2013)
G. Bussi, D. Donadio, M. Parrinello, Canonical sampling through velocity rescaling. J. Chem. Phys. 126 (2007)
H.J. Berendsen, J.V. Postma, W.F. Van Gunsteren, A. DiNola, J.R. Haak, Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984)
Z. Li, A molecular arm: the molecular bending-unbending mechanism of integrin. bioRxiv (2023). https://doi.org/10.1101/2023.04.10.536312
E. Puklin-Faucher, M. Gao, K. Schulten, V. Vogel, How the headpiece hinge angle is opened: new insights into the dynamics of integrin activation. J. Cell Biol. 175, 349–360 (2006)
Acknowledgements
This work is supported by grants from the National Natural Science Foundation of China (12272216 and 12172204) and the Natural Science Foundation of Shanghai (22ZR1423500).
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wang, K., Li, Z. Steered molecular dynamics simulation of force triggering the integrin αIIbβ3 extension via its ligand. Eur. Phys. J. Spec. Top. 232, 2773–2781 (2023). https://doi.org/10.1140/epjs/s11734-023-00965-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1140/epjs/s11734-023-00965-8