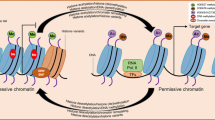
Abstract—The transition of embryos in air-dried seeds from a state of dormancy to a state with highly active metabolism during germination is accompanied by significant changes in both spatial and temporal patterns of gene expression and is controlled by multilevel regulatory networks. The character and degree of acetylation of chromatin proteins depend on the transcriptional activity of chromatin and are also associated with DNA replication and the cell cycle. Obtaining a complete picture of the involvement of histone modification in seed germination will be useful for increasing crop yields, as a way to assess the quality and viability of seeds before sowing, and will also allow the development of methods for managing plant genetic potential.

Similar content being viewed by others
REFERENCES
Alinsug, M.V., Chen, F.F., Luo, M., et al., Subcellular localization of class ii hdas in Arabidopsis thaliana: Nucleocytoplasmic shuttling of HDA15 is driven by light, PLoS One, 2012, vol. 7, p. e30846.
Azad, G.K., Swagatika, S., Kumawat, M., et al., Modifying chromatin by histone tail clipping, J. Mol. Biol., 2018, vol. 430, no. 18, pp. 3051–3067.
Boycheva, I., Vassileva, V., and Iantcheva, A., Histone acetyltransferases in plant development and plasticity, Curr. Genomics, 2014, vol. 15, no. 1, pp. 28–37.
Brownell, J.E. and Allis, C.D., Special hats for special occasions: Linking histone acetylation to chromatin assembly and gene activation, Curr. Opin. Genet. Dev., 1996, vol. 6, no. 2, pp. 176–184.
Carrera-Castaño, G., Calleja-Cabrera, J., Pernas, M., et al., An updated overview on the regulation of seed germination, Plants, 2020, vol. 9, no. 6, p. 703.
Chen, Z.J. and Tian, L., Roles of dynamic and reversible histone acetylation in plant development and polyploidy, Biochim. Biophys. Acta, 2007, vol. 1769, nos. 5–6, pp. 295–307.
Chen, W.Q., Li, D.X., Zhao, F., et al., One additional histone deacetylase and 2 histone acetyltransferases are involved in cellular patterning of Arabidopsis root epidermis, Plant Signaling Behav., 2016, vol. 11, p. e1131373.
Chhun, T., Chong, S.Y., Park, B.S., et al., HSI2 repressor recruits MED13 and hda6 to down-regulate seed maturation gene expression directly during Arabidopsis early seedling growth, Plant Cell Physiol., 2016, vol. 57, pp. 1689–1706.
Cimini, D., Mattiuzzo, M., Torosantucci, L., and Degrassi, F., Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects, Mol. Biol. Cell, 2003, vol. 14, no. 9, pp. 3821–3833.
Danovich, K.N., Sobolev, A.M., Zhdanova, L.P., et al., Fiziologiya semyan (Physiology of Seeds), Moscow: Nauka, 1982.
Davie, J.R., Inhibition of histone deacetylase activity by butyrate, J. Nutr., 2003, vol. 133, no. 7, pp. 2485S–2493S.
Dokmanovic, M. and Marks, P.A., Prospects: Histone deacetylase inhibitors, J. Cell. Biochem., 2005, vol. 96, no. 2, pp. 293–304.
Du, Z., Li, H., Wei, Q., et al., Genome-wide analysis of histone modifications: H3K4me2, H3K4me3, H3K9ac, and H3K27ac in Oryza sativa L. Japonica, Mol. Plant, 2013, vol. 6, no. 5, pp. 1463–1472.
Feitoza, L., Costa, L., and Guerra, M., Condensation patterns of prophase/prometaphase chromosome are correlated with H4K5 histone acetylation and genomic DNA contents in plants, PLoS One, 2017, vol. 12, no. 8, p. e0183341.
Fina, J.P., Masotti, F., Rius, S.P., et al., HAC1 and HAF1 histone acetyltransferases have different roles in UV-B responses in Arabidopsis, Front. Plant Sci., 2017, vol. 8, p. 1179.
Fuchs, J., Demidov, D., Houben, A., and Schubert, I., Chromosomal histone modification patterns—From conservation to diversity, Trends Plant Sci., 2006, vol. 11, pp. 199–208.
Gan, L., Wei, Z., Yang, Z., et al., Updated mechanisms of GCN5—The monkey king of the plant kingdom in plant development and resistance to abiotic stresses, Cells, 2021, vol. 10, no. 5, p. 979.
Garcia-Ramirez, M., Rocchini, C., and Ausio, J., Modulation of chromatin folding by histone acetylation, J. Biol. Chem., 1995, vol. 270, no. 30, pp. 17923–17928.
Gong, F., Chiu, L.Y., and Miller, K.M., Signaling to genome maintenance and cancer, PLoS Genet., 2016, vol. 12, no. 9, p. e1006272.
Haigney, A., Ricketts, M.D., and Marmorstein, R., Dissecting the molecular roles of histone chaperones in histone acetylation by type B histone acetyltransferases (HAT-B), J. Biol. Chem., 2015, vol. 290, no. 51, pp. 30648–30657.
Hartl, M., Fussl, M., Boersema, P.J., et al., Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis, Mol. Syst. Biol., 2017, vol. 13, no. 10, p. 949.
Hollender, C. and Liu, Z., Histone deacetylase genes in Arabidopsis development, J. Integr. Plant Biol., 2008, vol. 50, pp. 875–885.
Hong, L., Schroth, G.P., Matthews, H.P., et al., Studies of the dna binding properties of histone H4 amino terminus, J. Biol. Chem., 1993, vol. 268, no. 1, pp. 305–314.
Hou, H., Wang, P., Zhang, H., et al., Histone acetylation is involved in gibberellin-regulated sodCp gene expression in maize aleurone layers, Plant Cell Phys., 2015, vol. 56, no. 11, pp. 2139–2149.
Hu, Y., Lu, Y., Zhao, Y., and Zhou, D.X., Histone acetylation dynamics integrates metabolic activity to regulate plant response to stress, Front. Plant Sci., 2019, vol. 10, p. 1236.
Ivanov, V.B., Dobrochaev, A.E., and Baskin, T.I., What the distribution of cell lengths in the root meristem does and does not reveal about cell division, J. Plant Growth Regul., 2002, vol. 21, no. 1, pp. 60–67.
Jasencakova, Z., Meister, A., Walter, J., et al., Histone H4 acetylation of euchromatin and heterochromatin is cell cycle dependent and correlated with replication rather than with transcription, Plant Cell, 2000, vol. 12, pp. 2087–2100.
Jasencakova, Z., Meister, A., and Schubert, I., Chromatin organization and its relation to replication and histone acetylation during the cell cycle in barley, Chromosoma, 2001, vol. 110, no. 2, pp. 83–92.
Kim, S., Sophie, J.M., Piquerez, J.S., et al., GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5' and 3' ends of its target genes, Nucleic Acids Res., 2020, vol. 48, no. 11, pp. 5953–5966.
Kolle, D., Sarg, B., Lindner, H., and Loidl, P., Substrate and sequential site specificity of cytoplasmic histone acetyltransferases of maize and rat liver, FEBS Lett., 1998, vol. 421, no. 2, pp. 109–114.
Kouzarides, T., Chromatin modifications and their function, Cell, 2007, vol. 128, no. 4, pp. 693–705.
Kumar, V., Thakur, J.K., and Prasad, M., Histone acetylation dynamics regulating plant development and stress responses, Cell. Mol. Life Sci., 2021, vol. 78, no. 10, pp. 4467–4486.
Lechner, T., Lusser, A., Pipal, A., et al., RPD3-type histone deacetylases in maize embryos, Biochemistry, 2000, vol. 39, no. 7, pp. 1683–1692.
Lee, D.Y., Hayes, J.J., Pruss, D., Wolffe, A.P., et al., A positive role for histone acetylation in transcription factor access to nucleosomal DNA, Cell, 1993, vol. 72, no. 1, pp. 73–84.
Li, H., Torres-Garcia, J., Latrasse, D., et al., Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2–OXIDASE2 expression to control Arabidopsis root meristem cell number, Plant Cell, 2017, vol. 29, no. 9, pp. 2183–2196.
Li, Y., Butenko, Y., and Grafi, G., Histone deacetylation is required for progression through mitosis in tobacco cells, Plant J., 2005, vol. 41, no. 3, pp. 346–352.
Liu, C., Lu, F., Cui, X., and Cao, X., Histone methylation in higher plants, Annu. Rev. Plant Biol., 2010, vol. 61, pp. 395–420.
Liu, X., Chen, C.-Y., Wang, K.-C., et al., PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings, Plant Cell, 2013, vol. 25, no. 4, pp. 1258–1273.
Liu, X., Yang, S., Yu, C.W., et al., Histone acetylation and plant development, in Enzymes, Lin, C. and Luan, Sh., Eds., Burlington: Academic, 2016, vol. 40, pp. 173–199.
Loidl, P., Towards an understanding of the biological function of histone acetylation, FEBS Lett., 1988, vol. 227, no. 2, pp. 91–95.
Loidl, P., Histone acetylation: Facts and questions, Chromosoma, 1994, vol. 103, no. 7, pp. 441–449.
Lu, L., Chen, X., Sanders, D., et al., High-resolution mapping of H4K16 and H3K23 acetylation reveals conserved and unique distribution patterns in Arabidopsis and rice, Epigenetics, 2015, vol. 10, no. 11, pp. 1044–1053.
Luján-Soto, E.V. and Dinkova, T.D., Time to wake up: Epigenetic and small-RNA-mediated regulation during seed germination, Plants, 2021, vol. 10, no. 2, p. 236.
Mahrez, W., Arellano, M.S., Moreno-Romero, J., et al., H3K36ac is an evolutionary conserved plant histone modification that marks active genes, Plant Physiol., 2016, vol. 170, no. 3, pp. 1566–1577.
Mandal, P., Verma, N., Azad, G.K., et al., Epigenetics: Role of histone proteases in cellular functions and diseases, in Molecular Mechanisms and Physiology of Disease: Implications for Epigenetics and Health, Maulik, N. and Karagiannis, T., Eds., New York: Springer, 2014, pp. 113–125.
Mariño-Ramírez, L., Kann, M.G., Shoemaker, B.A., and Landsman, D., Histone structure and nucleosome stability, Expert Rev. Proteomics, 2005, vol. 2, no. 5, pp. 719–729.
Martin, B.J.E., Brind’Amour, J., Kuzmin, A., et al., Transcription shapes genome-wide histone acetylation patterns, Nat. Commun., 2021, vol. 12, no. 1, p. 210.
Martínez, Ó., Arjones, V., and González, M.V., Histone deacetylase inhibitors increase the embryogenic potential and alter the expression of embryogenesis-related and HDAC-encoding genes in grapevine (Vitis vinifera L., cv. Mencía), Plants, 2021, vol. 10, no. 6, p. 1164.
Musselman, C.A., Lalonde, M.E., Cote, J., and Kutateladze, T.G., Perceiving the epigenetic landscape through histone readers, Nat. Struct. Mol. Biol., 2012, vol. 19, no. 12, pp. 1218–1227.
Nguyen, H.N., Kim, J.H., Jeong, C.Y., et al., Inhibition of histone deacetylation alters Arabidopsis root growth in response to auxin via PIN1 degradation, Plant Cell Rep., 2013, vol. 32, no. 10, pp. 1625–1636.
Nieuwland, J., Stamm, P., Wen, B., et al., Re-induction of the cell cycle in the Arabidopsis post-embryonic root meristem is ABA-insensitive, GA-dependent and repressed by KRP6, Sci. Rep., 2016, vol. 6, p. 23586.
Nonogaki, H., Bassel, G.W., and Bewley, J.D., Germination—Still a mystery, Plant Sci., 2010, vol. 179, no. 6, pp. 574–581.
Nonogaki, H., Seed dormancy and germination–Emerging mechanisms and new hypotheses, Front. Plant Sci., 2014, vol. 5, p. 233.
Pandey, R., Muller, A., Napoli, C.A., et al., Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes, Nucleic Acids Res., 2002, vol. 30, no. 23, pp. 5036–5055.
Parthun, M.R., Widom, J., and Gottschling, D.E., The major cytoplasmic histone acetyltransferase in yeast: Links to chromatin replication and histone metabolism, Cell, 1996, vol. 87, no. 1, pp. 85–94.
Roth, S.Y., Denu, J.M., and Allis, C.D., Histone acetyltransferases, Annu. Rev. Biochem., 2001, vol. 70, pp. 81–120.
Sadoul, K., Boyault, C., Pabion, M., and Khochbin, S., Regulation of protein turnover by acetyltransferases and deacetylases, Biochimie, 2008, vol. 90, pp. 306–312.
Servet, C., Conde e Silva, N., and Zhou, D.-X., Histone acetyltransferase ATGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis, Mol. Plant, 2010, vol. 3, no. 4, pp. 670–677.
Smolikova, G., Strygina, K., Krylova, E., et al., Transition from seeds to seedlings: Hormonal and epigenetic aspects, Plants, 2021, vol. 10, no. 9, p. 1884.
Spencer, V.A. and Davie, J.R., Role of covalent modifications of histones on regulating gene expression, Gene, 1999, vol. 240, no. 1, pp. 1–12.
Tanaka, M., Kikuchi, A., and Kamada, H., The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination, Plant Physiol., 2008, vol. 146, no. 1, pp. 149–161.
Vafina, G.H. and Ivanov, R.S., Localization of Arg-X proteolysis in the supramolecular structures of cell nuclei during the induction of growth in mature wheat germs, Indian J. Plant Physiol., 2016, vol. 21, no. 3, pp. 370–373.
Vafina, G.H., Ivanov, R.S., and Ivanova, E.A., Analysis of Arg-X proteolytic activity in the supramolecular structures of cell nuclei influenced by inhibitor deacetylation of proteins during the germination of wheat, Indian J. Plant Physiol., 2017, vol. 3, pp. 358–364.
Vafina, G.H., Ivanov, R.S., and Ivanova, E.A., Changes of Arg-X proteolysis localization under conditions of deacetylation inhibition of nuclear proteins in spring and winter wheat seedlings, Acta Physiol. Plant., 2018, vol. 40, p. 78.
Vafina, G.H., Ivanov, R.S., and Kalashnik, N., Features of the formation of Arg-X proteolytic system of cellular nuclei during germination of wheat seeds, Bulg. J. Agric. Sci., 2020, vol. 26, no. 6, pp. 1158–1165. www.agrojournal.org/26/06–08.html.
Vettese-Dadey, M., Grant, P.A., Hebbes, T.R., et al., Acetylation of histone H4 plays a primary role in enhancing transcription factor binding to nucleosomal DNA in vitro, EMBO J., 1996, vol. 15, no. 10, pp. 2508–2518.
Wako, T., Fukuda, M., Furushima-Shimogawara, R., et al., Cell cycle-dependent and lysine residue-specific dynamic changes of histone H4 acetylation in barley, Plant Mol. Biol., 2002, vol. 49, no. 6, pp. 645–653.
Wang, Z., Cao, H., Sun, Y., et al., Arabidopsis paired amphipathic helix proteins SNL1 and SNL2 redundantly regulate primary seed dormancy via abscisic acid–ethylene antagonism mediated by histone deacetylation, Plant Cell, 2013, vol. 25, no. 1, pp. 149–166.
Wang, Z., Chen, F., Li, X., et al., Arabidopsis seed germination speed is controlled by SNL histone deacetylase-binding factor-mediated regulation of AUX1, Nat. Commun., 2016, vol. 7, p. 13412.
Wolny, E., Braszewska-Zalewska, A., Kroczek, D., and Hasterok, R., Histone H3 and H4 acetylation patterns are more dynamic than those of DNA methylation in Brachypodium distachyon embryos during seed maturation and germination, Protoplasma, 2017, vol. 254, pp. 2045–2052.
Xu, Q., Liu, Q., Chen, Z., et al., Histone deacetylases control lysine acetylation of ribosomal proteins in rice, Nucleic Acids Res., 2021, vol. 49, no. 8, pp. 4613–4628.
Xue, C., Liu, S., Chen, C., et al., Global proteome analysis links lysine acetylation to diverse functions in Oryza sativa, Proteomics, 2018, vol. 18, no. 1, p. 1700036.
Yadav, SP. and Das, H. K., Discontinuous incorporation of amino acids in embryo proteins of wheat during germination, Dev. Biol., 1974, vol. 36, no. 1, pp. 183–186.
Yang, W., Chen, Z., Huang, Y., et al., Powerdress as the novel regulator enhances Arabidopsis seeds germination tolerance to high temperature stress by histone modification of SOM locus, Plant Sci., 2019, vol. 284, pp. 91–98.
Yano, R., Takebayashi, Y., Nambara, E., et al., Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana, Plant J., 2013, vol. 74, no. 5, pp. 815–828.
Yruela, I., Moreno-Yruela, C., and Olsen, C.A., Zn2+-dependent histone deacetylases in plants: Structure and evolution, Trends Plant Sci., 2021, vol. 26, no. 7, pp. 741–757.
van Zanten, M., Koini, M.A., Geyer, R., et al., Seed maturation in Arabidopsis thaliana is characterized by nuclear size reduction and increased chromatin condensation, Proc. Natl. Acad. Sci. U. S. A., 2011, vol. 108, no. 50, pp. 20219–20224.
van Zanten, M., Zöll, C., Wang, Z., et al., Histone deacetylase 9 represses seedling traits in Arabidopsis thaliana dry seeds, Plant J., 2014, vol. 80, no. 3, pp. 475–488.
Zhang, H. and Ogas, J., An epigenetic perspective on developmental regulation of seed genes, Mol. Plant, 2009, vol. 2, no. 4, pp. 610–627.
Zhang, Q., Wang, P., Hou, H., et al., Histone acetylation and reactive oxygen species are involved in the preprophase arrest induced by sodium butyrate in maize roots, Protoplasma, 2017, vol. 254, pp. 167–179.
Zhao, L., Peng, T., Chen, C.-Y., et al., HY5 interacts with the histone deacetylase HDA15 to repress hypocotyl cell elongation in photomorphogenesis, Plant Physiol., 2019, vol. 180, no. 3, pp. 1450–1466.
Zhou, Y., Tan, B., Luo, M., et al., HISTONE DEACETYLASE 19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings, Plant Cell, 2013, vol. 25, no. 1, pp. 134–148.
ACKNOWLEDGMENTS
We express our deep gratitude to Professor R.N. Churaev for valuable comments and support.
Funding
This study was carried out within the state task of the RF Ministry of Science and Higher Education no. 075-00326-19-00 on topic AAAA-A18-118022190099-6 and no. 075-03-2021-607 on topic no. 122031100163-4.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest. The authors declare that they have no conflicts of interest.
Statement of the welfare of humans or animals. The article does not contain any studies involving humans or animals in experiments performed by any of the authors.
Rights and permissions
About this article
Cite this article
Vafina, G.H., Stupak, E.E. On the Biological Role of Histone Acetylation/Deacetylation in the Process of Plant Germination. Biol Bull Rev 13, 140–147 (2023). https://doi.org/10.1134/S2079086423020093
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079086423020093