Abstract—
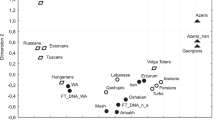
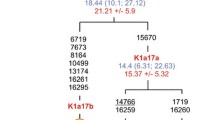
Data on the variability of the nucleotide sequences of whole mitochondrial genomes in populations of Russians, Poles, and Serbs, which represent, respectively, the groups of East, West, and South Slavs, are reviewed in comparison with similar data for other European peoples. It was shown that the results of the analysis of the variability of whole mitogenomes make it possible to substantially detail the ideas about the structure and diversity of mitochondrial gene pools obtained earlier with other approaches, e.g., analysis of the distribution of haplotypes of mtDNA major noncoding region or the frequencies of mitochondrial haplogroups. It has been demonstrated that Bayesian analysis of large data sets on the variability of whole mitogenomes in the Slavs enabled the identification of between-regional differences in the dynamics of the effective population size over time for Europe. The use of a phylogeographic analysis of modern and paleogenomic data on the variability of whole mitogenomes makes it possible to define unique phylogenetic mtDNA clusters specific to certain ethnic groups and their communities. Their common genetic origin, which dates back to the Bronze and Iron Ages, is demonstrated on the example of three ethnic groups of Slavs.





Similar content being viewed by others
REFERENCES
Alekseeva, T.I., Etnogenez vostochnykh slavyan po dannym antropologii (Ethnogenesis of Eastern Slavs According to Anthropological Data), Moscow: Mosk. Gos. Univ., 1973.
Alekseeva, T.I. and Alekseev, V.P., Anthropological view on the origin of Slavs, Priroda (Moscow), 1989, no. 1, pp. 60–62.
Allentoft, M.E., Sikora, M., Sjögren, K.G., et al., Population genomics of Bronze Age Eurasia, Nature, 2015, vol. 522, pp. 167–172.
Aris-Brosou, S. and Excoffier, L., The impact of population expansion and mutation rate heterogeneity on DNA sequence polymorphism, Mol. Biol. Evol., 1996, vol. 13, pp. 494–504.
Balanovsky, O., Rootsi, S., Pshenichnov, A., et al., Two sources of the Russian patrilineal heritage in their Eurasian context, Am. J. Hum. Genet., 2008, vol. 82, pp. 236–250.
Batini, C., Hallast, P., Vågene, Å.J., et al., Population resequencing of European mitochondrial genomes highlights sex-bias in Bronze Age demographic expansions, Sci. Rep., 2017, vol. 7, p. e12086.
Behar, D.M., van Oven, M., Rosset, S., et al., A “Copernican” reassessment of the human mitochondrial DNA tree from its root, Am. J. Hum. Genet., 2012, vol. 90, pp. 675–684.
Busby, G.B.J., Hellenthal, G., Montinaro, F., et al., The role of recent admixture in forming the contemporary West Eurasian genomic landscape, Curr. Biol., 2015, vol. 25, p. 2878.
Comas, D., Calafell, F., Mateu, E., et al., Geographic variation in human mitochondrial DNA control region sequence: the population history of Turkey and its relationship to the European populations, Mol. Biol. Evol., 1996, vol. 13, pp. 1067–1077.
Costa, M.D., Pereira, J.B., Pala, M., et al., A substantial prehistoric European ancestry amongst Ashkenazi maternal lineages, Nat. Commun., 2013, vol. 4, p. e2543.
Csáky, V., Gerber, D., Koncz, I., et al., Genetic insights into the social organisation of the Avar period elite in the 7th century AD Carpathian Basin, Sci. Rep., 2020, vol. 10, p. e948.
Csákyová, V., Szécsényi-Nagy, A., Csősz, A., et al., Maternal genetic composition of a medieval population from a Hungarian-Slavic contact zone in Central Europe, PLoS One, 2016, vol. 11, p. e0151206.
Csősz, A., Szécsényi-Nagy, A., Csákyová, V., et al., Maternal genetic ancestry and legacy of 10th century AD Hungarians, Sci. Rep., 2016, vol. 6, p. e33446.
Davidović, S., Malyarchuk, B., Aleksić, J., et al., Mitochondrial DNA perspective of Serbian genetic diversity, Am. J. Phys. Anthropol., 2015, vol. 156, pp. 449–465.
Davidović, S., Malyarchuk, B., Aleksić, J., et al., Mitochondrial super-haplogroup U diversity in Serbians, Ann. Hum. Biol., 2017, vol. 44, pp. 408–418.
Davidović, S., Malyarchuk, B., Grzybowski, T., et al., Complete mitogenome data for the Serbian population: the contribution to high quality forensic databases, Int. J. Legal Med., 2020, vol. 134, pp. 1581-1590.
Denaro, M., Blanc, H., Johnson, M.J., et al., Ethnic variation in Hpa I endonuclease cleavage patterns of human mitochondrial DNA, Proc. Natl. Acad. Sci. U.S.A., 1981, vol. 78, pp. 5768–5772.
Derenko, M., Malyarchuk, B., Bahmanimehr, A., et al., Complete mitochondrial DNA diversity in Iranians, PLoS One, 2013, vol. 8, p. e80673.
Derenko, M., Denisova, G., Malyarchuk, B., et al., Mitogenomic diversity and differentiation of the Buryats, J. Hum. Genet., 2018, vol. 63, pp. 71–81.
Derenko, M., Denisova, G., Malyarchuk, B., et al., Insights into matrilineal genetic structure, differentiation and ancestry of Armenians based on complete mitogenome data, Mol. Genet. Genomics, 2019, vol. 294, pp. 1547–1559.
Drummond, A.J., Nicholls, G.K., Rodrigo, A.G., and Solomon, W., Estimating mutation parameters, population history and genealogy simultaneously from temporally spaced sequence data, Genetics, 2002, vol. 161, pp. 1307–1320.
Drummond, A.J., Rambaut, A., Shapiro, B., and Pybus, O.G., Bayesian coalescent inference of past population dynamics from molecular sequences, Mol. Biol. Evol., 2005, vol. 22, pp. 1185–1192.
Egyed, B., Brandstätter, A., Irwin, J.A., et al., Mitochondrial control region sequence variations in the Hungarian population: analysis of population samples from Hungary and from Transylvania (Romania), Forensic Sci. Int. Genet., 2007, vol. 1, pp. 158–162.
Fraumene, C., Belle, E.M., Castri, L., et al., High resolution analysis and phylogenetic network construction using complete mtDNA sequences in Sardinian genetic isolates, Mol. Biol. Evol., 2006, vol. 23, pp. 2101–2111.
Grzybowski, T., Malyarchuk, B.A., Derenko, M.V., et al., Complex interactions of the Eastern and Western Slavic populations with other European groups as revealed by mitochondrial DNA analysis, Forensic Sci. Int. Genet., 2007, vol. 1, pp. 141–147.
Haak, W., Lazarids, I., Patterson, N., et al., Massive migration from the steppe was a source for Indo-European languages in Europe, Nature, 2015, vol. 522, pp. 207–211.
Hellenthal, G., Busby, G.B.J., Band, G., et al., A genetic atlas of human admixture history, Science, 2014, vol. 343, pp. 747–751.
Hofmanová, Z., Kreutzer, S., Hellenthal, G., et al., Early farmers from across Europe directly descended from Neolithic Aegeans, Proc. Natl. Acad. Sci. U.S.A., 2016, vol. 113, pp. 6886–6891.
Gimbutas, M., The Prehistory of Eastern Europe, Part 1: Mesolithic, Neolithic and Copper Age Cultures in Russia and the Baltic Area, Cambridge: Peabody Mus., 1956.
Järve, M., Saag, L., Scheib, C.L., et al., Shifts in the genetic landscape of the Western Eurasian steppe associated with the beginning and end of the Scythian dominance, Curr. Biol., 2019, vol. 29, pp. 2430–2441.
Johnson, M.J., Wallace, D.C., Ferris, S.D., et al., Radiation of human mitochondria DNA types analyzed by restriction endonuclease cleavage patterns, J. Mol. Evol., 1983, vol. 19, pp. 255–271.
Juras, A., Dabert, M., Kushniarevich, A., et al., Ancient DNA reveals matrilineal continuity in present-day Poland over the last two millennia, PLoS One, 2014, vol. 9, p. e110839.
Just, R.S., Scheible, M.K., Fast, S.A., et al., Full mtGenome reference data: development and characterization of 588 forensic-quality haplotypes representing three U.S. populations, Forensic Sci. Int. Genet., 2015, vol. 14, pp. 141–155.
Karachanak, S., Carossa, V., Nesheva, D., et al., Bulgarians vs the other European populations: a mitochondrial DNA perspective, Int. J. Legal Med., 2012, vol. 126, pp. 497–503.
Karmin, M., Saag, L., Vicente, M., et al., A recent bottleneck of Y chromosome diversity coincides with a global change in culture, Genome Res., 2015, vol. 25, pp. 459–466.
Khrunin, A.V., Khokhrin, D.V., Filippova, I.N., et al., A genome-wide analysis of populations from European Russia reveals a new pole of genetic diversity in northern Europe, PLoS One, 2013, vol. 8, p. e58552.
Kushniarevich, A., Utevska, O., Chuhryaeva, M., et al., Genetic heritage of the Balto-Slavic speaking populations: a synthesis of autosomal, mitochondrial and Y‑chromosomal data, PLoS One, 2015, vol. 10, p. e0135820.
Ler-Splavinskii, T., Origin of the Slavs, Vopr. Yazykozn., 1960, no. 4, pp. 20–30.
Li, S., Besenbacher, S., Li, Y., et al., Variation and association to diabetes in 2000 full mtDNA sequences mined from an exome study in a Danish population, Eur. J. Hum. Genet., 2014, vol. 22, pp. 1040–1045.
Librado, P. and Rozas, J., DnaSP v5: a software for comprehensive analysis of DNA polymorphism data, Bioinformatics, 2009, vol. 25, pp. 1451–1452.
Lippold, S., Xu, H., Ko, A., et al., Human paternal and maternal demographic histories: insights from high-resolution Y chromosome and mtDNA sequences, Invest. Genet., 2014, vol. 5, p. e13.
Lipson, M., Szécsényi-Nagy, A., Mallick, S., et al., Parallel palaeogenomic transects reveal complex genetic history of early European farmers, Nature, 2017, vol. 551, pp. 368–372.
Malyarchuk, B.A., Differentiation and genetic position of slavs among Eurasian ethnic groups as inferred from variation in mitochondrial DNA, Russ. J. Genet., 2001, vol. 37, no. 12, pp. 1437–1443.
Malyarchuk, B.A., Sources of the mitochondrial gene pool of Russians by the results of analysis of modern and paleogenomic data, Vavilovsk. Zh. Genet. Sel., 2019, vol. 23, no. 5, pp. 588–593.
Malyarchuk, B.A. and Derenko, M.V., Mitochondrial DNA variability in Russians and Ukrainians: implication to the origin of the Eastern Slavs, Ann. Hum. Genet., 2001, vol. 65, pp. 63–78.
Malyarchuk, B.A., Grzybowski, T., Derenko, M.V., et al., Mitochondrial DNA variability in Bosnians and Slovenians, Ann. Hum. Genet., 2003, vol. 67, pp. 412–425.
Malyarchuk, B., Derenko, M., Grzybowski, T., et al., Differentiation of mitochondrial DNA and Y chromosomes in Russian populations, Hum. Biol., 2004, vol. 76, pp. 877–900.
Malyarchuk, B.A., Grzybowski, T., Derenko, M., et al., Mitochondrial DNA phylogeny in Eastern and Western Slavs, Mol. Biol. Evol., 2008, vol. 25, pp. 1651–1658.
Malyarchuk, B., Derenko, M., Denisova, G., and Kravtsova, O., Mitogenomic diversity in Tatars from the Volga-Ural region of Russia, Mol. Biol. Evol., 2010, vol. 27, pp. 2220–2226.
Malyarchuk, B.A., Derenko, M.V., and Litvinov, A.N., The macrohaplogroup U structure in Russians, Russ. J. Genet., 2017a, vol. 53, no. 4, pp. 498–503.
Malyarchuk, B., Litvinov, A., Derenko, M., et al., Mitogenomic diversity in Russians and Poles, Forensic Sci. Int. Genet., 2017b, vol. 30, pp. 51–56.
Malyarchuk, B., Derenko, M., Denisova, G., et al., Whole mitochondrial genome diversity in two Hungarian populations, Mol. Gen. Genomics, 2018, vol. 293, pp. 1255–1263.
Malyarchuk, B.A., Litvinov, A.N., and Derenko, M.V., Structure and forming of mitochondrial gene pool of Russian population of Eastern Europe, Russ. J. Genet., 2019, vol. 55, no. 5, pp. 622–629.
Margaryan, A., Lawson, D., Sikora, M., et al., Population genomics of the Viking world, Nature, 2020, vol. 585, pp. 390–396. https://doi.org/10.1038/s41586-020-2688-8
Mielnik-Sikorska, M., Daca, P., Malyarchuk, B., et al., The history of Slavs inferred from complete mitochondrial genome sequences, PLoS One, 2013, vol. 8, p. e54360.
Morozova, I., Evsyukov, A., Kon’kov, A., et al., Russian ethnic history inferred from mitochondrial DNA diversity, Am. J. Phys. Anthropol., 2012, vol. 147, pp. 341–351.
Müller, J., Rassmann, K., and Videiko, M., Trypillia Mega-Sites and European Prehistory: 4100-3400 BCE, London: Routledge, 2016.
Neparáczki, E., Juhász, Z., Pamjav, H., et al., Genetic structure of the early Hungarian conquerors inferred from mtDNA haplotypes and Y-chromosome haplogroups in a small cemetery, Mol. Genet. Genomics, 2017, vol. 292, pp. 201–214.
Olivieri, A., Sidore, C., Achilli, A., et al., Mitogenome diversity in Sardinians: a genetic window onto an Island’s past, Mol. Biol. Evol., 2017, vol. 34, pp. 1230–1239.
Översti, S., Onkamo, P., Stoljarova, M., et al., Identification and analysis of mtDNA genomes attributed to Finns reveal long-stagnant demographic trends obscured in the total diversity, Sci. Rep., 2017, vol. 7, p. e6193.
Pankratov, V., Litvinov, S., Kassian, A., et al., East Eurasian ancestry in the middle of Europe: genetic footprints of steppe nomads in the genomes of Belarusian Lipka Tatars, Sci. Rep., 2016, vol. 6, p. e30197.
Piotrowska-Nowak, A., Elson, J.L., Sobczyk-Kopciol, A., et al., New mtDNA association model, MutPred variant load, suggests individuals with multiple mildly deleterious mtDNA variants are more likely to suffer from atherosclerosis, Front. Genet., 2019a, vol. 9, p. 702.
Piotrowska-Nowak, A., Kosior-Jarecka, E., Schab, A., et al., Investigation of whole mitochondrial genome variation in normal tension glaucoma, Exp. Eye Res., 2019b, vol. 178, pp. 186–197.
Pereira, J.B., Costa, M.D., Vieira, D., et al., Reconciling evidence from ancient and contemporary genomes: a major source for the European Neolithic within Mediterranean Europe, Proc. Biol. Sci., 2017, vol. 284, p. e20161976.
Ralph, P. and Coop, G., The geography of recent genetic ancestry across Europe, PLoS Biol., 2013, vol. 11, p. e1001555.
Rascovan, N., Sjögren, K.-G., Kristiansen, K., et al., Emergence and spread of basal lineages of Yersinia pestis during the Neolithic decline, Cell, 2019, vol. 176, pp. 295–305.
Raule, N., Sevini, F., Li, S., et al., The co-occurrence of mtDNA mutations on different oxidative phosphorylation subunits, not detected by haplogroup analysis, affects human longevity and is population specific, Aging Cell, 2014, vol. 13, pp. 401–407.
Ray, N., Currat, M., and Excoffier, L., Intra-deme molecular diversity in spatially expanding populations, Mol. Biol. Evol., 2003, vol. 20, pp. 76–86.
Rebała, K., Mikulich, A.I., Tsybovsky, I.S., et al., Y-STR variation among Slavs: evidence for the Slavic homeland in the middle Dnieper basin, J. Hum. Genet., 2007, vol. 52, pp. 406–414.
Rogers, A.R. and Harpending, H., Population growth makes waves in the distribution of pairwise genetic differences, Mol. Biol. Evol., 1992, vol. 9, pp. 552–569.
Sarac, J., Saric, T., Augustin, D.H., et al., Maternal genetic heritage of Southeastern Europe reveals a new Croatian isolate and a novel, local sub-branching in the X2 haplogroup, Ann. Hum. Genet., 2014, vol. 78, pp. 178–194.
Sedov, V.V., Proiskhozhdenie i rannyaya istoriya slavyan (Origin and Early History of the Slavs), Moscow: Nauka, 1979.
Sedov, V.V., Slavyane v rannem srednevekov’e (Slavs in the Early Middle Ages), Moscow: Nauka, 1995.
Sedov, V.V., Ethnogenesis of the early Slavs, Vestn. Ross. Akad. Nauk, 2003, vol. 73, pp. 594–605.
Shennan, S., Downey, S.S., Timpson, A., et al., Regional population collapse followed initial agriculture booms in mid-Holocene Europe, Nat. Commun., 2013, vol. 4, p. e2486.
Skonieczna, K., Malyarchuk, B., Jawień, A., et al., Mitogenomic differences between the normal and tumor cells of colorectal cancer patients, Hum. Mutat., 2018, vol. 39, pp. 691–701.
Soares, P., Ermini, L., Thomson, N., et al., Correcting for purifying selection: an improved human mitochondrial molecular clock, Am. J. Hum. Genet., 2009, vol. 84, pp. 740–759.
Stoljarova, M., King, J.L., Takahashi, M., et al., Whole mitochondrial genome genetic diversity in an Estonian population sample, Int. J. Leg. Med., 2016, vol. 130, pp. 67–71.
Tajima, F., Statistical method for testing the neutral mutation hypothesis by DNA polymorphism, Genetics, 1989, vol. 123, pp. 585–595.
Tajima, F., Statistical analysis of DNA polymorphism, Jpn. J. Genet., 1993, vol. 68, pp. 567–595.
Tömöry, G., Csányi, B., Bogácsi-Szabó, E., et al., Comparison of maternal lineage and biogeographic analyses of ancient and modern Hungarian populations, Am. J. Phys. Anthropol., 2007, vol. 134, pp. 354–368.
Woźniak, M., Malyarchuk, B., Derenko, M., et al., Similarities and distinctions in Y chromosome gene pool of Western Slavs, Am. J. Phys. Anthropol., 2010, vol. 142, pp. 540–548.
Zheng, H.-X., Yan, S., Qin, Z.-D., and Jin, L., mtDNA analysis of global populations support that major population expansions began before Neolithic Time, Sci. Rep., 2012, vol. 2, p. e745.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests. The authors declare that they have no conflicts of interest.
Statement on the welfare of humans or animals. This article does not contain any studies involving animals performed by any of the authors.
Additional information
Translated by A. Kazantseva
Rights and permissions
About this article
Cite this article
Malyarchuk, B.A., Derenko, M.V. Diversity and Structure of Mitochondrial Gene Pools of Slavs in the Ethnogenetic Aspect. Biol Bull Rev 11, 122–133 (2021). https://doi.org/10.1134/S2079086421020067
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S2079086421020067