Abstract
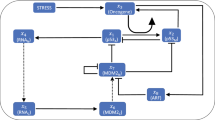
A hierarchy of minimal mathematical models of the dynamics of the p53-Mdm2- microRNA system has been developed. The models are based on differential equations with a time delay, describing complex interaction mechanisms in the signal system of the p53 protein. We consider two types of interaction of p53 with microRNAs: a positive direct connection and a positive feedback. The feedback of microRNA-p53 is due to a negative effect of the microRNA on the Mdm2 protein, which is a negative regulator of p53. To approximate the direct positive effect of p53 on the microRNAs, a linear function or a representation of the Goldbeter-Koshland type is used. A comparison of numerical solutions with medical and biological data of a number of specific p53-dependent microRNAs is made, which proves that the models and the numerical analysis results are adequate. Special attention is given to analysis of a positive feedback of p53 and microRNAs. The minimal models allow us to consider the most general regularities of the p53-dependent microRNAs. Within the framework of these mathematical models it is shown that it is possible to neglect the Mdm2-miRNA connection for at least some of the most studied microRNAs associated with a direct positive connection with p53. However, those of the microRNAs that are an important negative regulator of Mdm2, can have the most significant impact on the entire p53-Mdm2-microRNA system. Conditions are obtained to manifest the regulatory function of microRNAs with respect to p53. The results of the numerical experiments indicate that such microRNAs can be used as a factor of an anticancer therapy.
Similar content being viewed by others
References
Almazov, V.P., Kochetkov, D.V., and Chumakov, P.M., Use of p53 for Therapy of Human Cancer, Molec. Biol., 2007, vol. 41, no. 6, pp. 947-963.
Lane, D. and Levine, A., p53 Research: The Past Thirty Years and the Next Thirty Years, Cold Spring Harb. Perspect. Biol., 2010, vol. 2, no. 12, (a000893); DOI: 10.1101/cshperspect.a000893.
Geva-Zatorsky, N., Rosenfeld, N., Itzkovitz, Sh., et al., Oscillations and Variability in the p53 System, Molec. Syst. Biol., 2006, vol. 2, no. 1, (2006.0033); DOI: 10.1038/msb4100068.
Batchelor, E., Mock, C.S., Bhan, I., Loewer, A., and Lahav, G., Recurrent Initiation: A Mechanism for Triggering p53 Pulses in Response to DNA Damage, Mol. Cell., 2008, vol. 30, no. 3, pp. 277–289.
Toettcher, J.E., Mock, C., Batchelor, E., Loewer, A., and Lahav, G., A Synthetic-Natural Hybrid Oscillator in Human Cells, PNAS, 2010, vol. 107, no. 39, pp. 17047–17052.
Purvis, J.E., Karhohs, K.W., Mock, C., et al., p53 Dynamics Control Cell Fate, Science, 2012, vol. 336, iss. 6087, pp. 1440–1444.
Paek, A.L., Liu, J.C., Loewer, A., Forrester, W.C., and Lahav, G., Cell-to-Cell Variation in p53 Dynamics Leads to Fractional Killing, Cell, 2016, vol. 165, no. 3, pp. 631–642.
Goeman, F., Strano, S., and Blandino, G., MicroRNAs as Key Effectors in the p53 Network, in Int. Review of Cell and Molecular Biology, vol. 333, MiRNAs in Differentiation and Development, Galluzzi, L. and Vitale, I., Eds., Academic Press, 2017, pp. 51–91.
Hermeking, H., MicroRNAs in the p53 Network: Micromanagement of Tumor Suppression, Nature Reviews Cancer, 2012, vol. 12, no. 9, pp. 613–626.
Kolesnikov, N.N., Titov, S.E., Veryaskina, Yu.A., et al., MicroRNA, Evolution and Cancer, Tsitol., 2013, vol. 55, no. 3, pp. 159–164.
Rokavec, M., Li, H., Jiang, L., and Hermeking, H., The p53/microRNA Connection in Gastrointestinal Cancer, Clin. Exper. Gastroenterol., 2014, vol. 7, pp. 395–413.
Raver-Shapira, N., Marciano, E., Meiri, E., et al., Transcriptional Activation of miR-34a Contributes to p53-Mediated Apoptosis, Molec. Cell, 2007, vol. 26, pp. 731–743.
Bisio, A., Sanctis, V., Vescovo, V., et al., Identification of New p53 Target MicroRNAs by Bioinformatics and Functional Analysis, BMC Cancer, 2013, vol. 13, no. 552; DOI: 10.1186/1471-2407-13-552.
Zhang, J., Sun, Q., Zhang, Z., et al., Loss of MicroRNA-143/145 Disturbs Cellular Growth and Apoptosis of Human Epithelial Cancers by Impairing the Mdm2-p53 Feedback Loop, Oncogene, 2013, vol. 32, no. 1, pp. 61–69.
Fornari, F., Milazzo, M., Galassi, M., et al., p53/Mdm2 Feedback Loop Sustains miR-221 Expression and Dictates the Response to Anticancer Treatments in Hepatocellular Carcinoma, Molec. Cancer Res., 2014, vol. 12, no. 2, pp. 203–216.
Luo, Z., Cui, R., Tili, E., and Croce, C., Friend or Foe: MicroRNAs in the p53 Network, Cancer Lett., 2018, vol. 419, pp. 96–102.
Voropaeva, O.F., Shokin, Yu.I., Nepomnyashchikh, L.M., and Senchukova, S.R., Mathematical Simulation of p53-Mdm2 Protein Biological System Regulation, Bull. Exper. Biol. Medicine, 2014, vol. 157, no. 4, pp. 535–538.
Voropaeva, O.F., Senotrusova, S.D., and Shokin, Yu.I., Deregulation of p53-Dependent MicroRNAs: The Results of Mathematical Modeling, Math. Biol. Bioinform., 2017, vol. 12, no. 1, pp. 151–175.
Likhoshvai, V.A., Fadeev, S.I., Demidenko, G.V., and Matushkin, Yu.G., Modeling of Multistage Synthesis without Branching by an Equation with a Delay Argument, Sib. Zh. Ind. Mat., 2004, vol. 7, no. 1, pp. 73–94.
Tiana, G., Jensen, M.H., and Sneppen, K., Time Delay as a Key to Apoptosis Induction in the p53 Network, Eur. Phys. J. B., 2002, no. 29, pp. 135–140.
Voropaeva, O.F. and Senotrusova, S.D., The Passage from Delay Equation to ODE System in the Model of the Tumor Markers Network, Mat. Model., 2017, vol. 29, no. 9, pp. 135–154.
Voropaeva, O.F., Senotrusova, S.D., and Shokin, Y.I., Numerical Investigation of Diagnostic Properties of p53-Dependent MicroRNAs, RJNAMM, 2017, vol. 32, no. 3, pp. 203–213.
Chen, C.Y., Oliner, J.D., Zhan, Q., et al., Interactions between p53 and MDM2 in a Mammalian Cell Cycle Checkpoint Pathway, Proc. Natl. Acad. Sci. USA, 1994, vol. 91, no. 7, pp. 2684–2688.
Yang, R., Huang, B., Zhu, Y., et al., Variable Sensitivity to DNA Damaging Chemotherapeutic Modulated by Cell Type-Dependent Bimodal p53 Dynamics, bioRxiv, URL: https://www.biorxiv.org/content/early/2017/06/12/149013.
Iorio, M.V., Visone, R., Leva, G., et al., MicroRNA Signatures in Human Ovarian Cancer, Cancer Res., 2007, vol. 67, pp. 8699–8707.
Shulenina, L.V., Mikhailov, V.F., Ledin, E.V., Raeva, N.F., and Zasukhina, G.D., Evaluation of P53-Dependent System of Maintaining the Genome Stability by Content of MicroRNA and MRNA in Blood of Cancer Patients, Med. Radiol. Radiat. Saf., 2015, vol. 60, no. 1, pp. 5–14.
Yu, J., Baron V., Mercola, D., Mustelin, T., and Adamson, E.D., A Network of p73, p53 and Egr1 is Required for Efficient Apoptosis in Tumor Cells, Cell Death Diff., 2007, vol. 14, pp. 436–446.
Castro, R.E., Ferreira, D.M.S., Afonso, M.B., et al., miR-34a/SIRT1/p53 Is Suppressed by Ursodeoxycholic Acid in the Rat Liver and Activated by Disease Severity in Human Non-Alcoholic Fatty Liver Disease, J. Hepatol., 2013, vol. 58, iss. 1, pp. 119–125.
Kato, R., Mizuno, Sh., Kadowaki, M., et al., Sirt1 Expression Is Associated with CD31 Expression in Blood Cells from Patients with Chronic Obstructive Pulmonary Disease, Respir. Res., 2016, vol. 17, no. 139 (PMC5081972).
Baker, J.R., Vuppusetty, C., Colley, T., et al., Oxidative Stress Dependent MicroRNA-34a Activation via PI3K? Reduces the Expression of Sirtuin-1 and Sirtuin-6 in Epithelial Cells, Sci. Rep., 2016, vol. 6 (35871; PMC5073335).
Moore, R., Ooi, H.K., Kang, T., Bleris, L., and Ma, L., MiR-192-Mediated Positive Feedback Loop Controls the Robustness of Stress-Induced p53 Oscillations in Breast Cancer Cells, PLoS Comput. Biol., 2015, vol. 11, no. 12 (e1004653; PMC4671655).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Russian Text © The Author(s), 2019, published in Sibirskii Zhurnal Vychislitel’noi Matematiki, 2019, Vol. 22, No. 3, pp. 317–334.
Rights and permissions
About this article
Cite this article
Senotrusova, S.D., Voropaeva, O.F. Mathematical Modeling of a Positive Connection in the p53-microRNA Tumor Marker System. Numer. Analys. Appl. 12, 270–283 (2019). https://doi.org/10.1134/S1995423919030066
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1995423919030066