Abstract
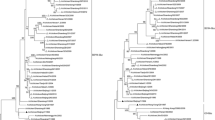
Influenza A virus has a wide natural areal among birds, mammals, and humans. One of the main regulatory adaptors of the virus host range is the major NP protein of the viral nucleocapsid. Phylogenetic analysis of the NP protein of different viruses has revealed the existence of two phylogenetic cohorts in human influenza virus population. Cohort I includes classical human viruses that caused epidemics in 1957, 1968, 1977. Cohort II includes the H1N1/2009pdm virus, which had a mixed avian–swine origin but caused global human pandemic. Also, the highly virulent H5N1 avian influenza virus emerged in 2021 and caused outbreaks of lethal infections in mammals including humans, appeared to have the NP gene of the second phylogenetic cohort and, therefore, by the type of adaptation to human is similar to the H1N1/2009pdm virus and seems to possess a high epidemic potential for humans. The data obtained shed light on pathways and dynamics of adaptation of avian influenza viruses to humans and propose phylogenetic algorithm for systemic monitoring of dangerous virus strains to predict epidemic harbingers and take immediate preventive measures.


Similar content being viewed by others
REFERENCES
Walker, P.J., Siddell, S.G., Lefkowitz, E.J., et al., Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022), Arch Virol., 2022, vol. 167, no. 11, pp. 2429–2440.
Zhirnov, O.P., The unique genome of the virus and alternative strategies for its realization, Acta Nat., 2023, vol. 15, no. 2, pp. 14–19.https://doi.org/10.32607/actanaturae.1190437538802
Zhirnov, O.P., The host origin of influenza A viruses can be assessed by the intracellular cleavage of the viral nucleocapsid protein, Arch Virol., 1988, vol. 99, nos. 3–4, pp. 277–284.
Mänz, B., Dornfeld, D., Götz, V., Zell, R., Zimmermann, P., Haller, O., Kochs, G., and Schwemmle, M., Pandemic influenza A viruses escape from restriction by human MxA through adaptive mutations in the nucleoprotein, PLoS Pathog., 2013, vol. 9, no. 3, p. e1003279. https://doi.org/10.1371/journal.ppat.100327923555271
Hall, J.S., Teslaa, J.L., Nashold, S.W., et al., Evolution of a reassortant North American gull influenza virus lineage: drift, shift and stability, Virol. J., 2013, vol. 10, p. 179.
Lycett, S.J., Duchatel, F., and Digard, P., A brief history of bird flu, Philos. Trans. R. Soc., B, 2019, vol. 374, no. 1775, p. 20180257.
WHO, Ongoing Avian Influenza Outbreaks in Animals Pose Risk to Humans, 2023. www.who.int/news/item/12-07-2023-ongoing-avian-influenza-outbreaks-in-animals-pose-risk-to-humans.
Haller, O. and Kochs, G., Mx genes: host determinants controlling influenza virus infection and trans-species transmission, Hum. Genet., 2020, vol. 139, nos. 6–7, pp. 695–705.
Peacock, T.P., Sheppard, C.M., Lister, M.G., et al., Mammalian ANP32A and ANP32B proteins drive differential polymerase adaptations in avian influenza virus, J. Virol., 2023, vol. 97, no. 5, p. e0021323.
Tome-Amat, J., Ramos, I., Amanor, F., Fernández-Sesma, A., and Ashour, J., Influenza A virus utilizes low-affinity, high-avidity interactions with the nuclear import machinery to ensure infection and immune evasion, J. Virol., 2018, vol. 93, no. 1, p. e01046-18.
Morris, A.K., Wang, Z., Ivey, A.L., Xie, Y., Hill, P.S., Schey, K.L., and Ren, Y., Cellular mRNA export factor UAP56 recognizes nucleic acid binding site of influenza virus NP protein, Biochem. Biophys. Res. Commun., 2020, vol. 525, no. 2, pp. 259–264.
York, I. and Donis, R.O., The 2009 pandemic influenza virus: where did it come from, where is it now, and where is it going?, Curr. Top. Microbiol. Immunol., 2013, vol. 370, pp. 241–257.
Gamarra-Toledo, V., Plaza, P.I., Inga, G., et al., First mass mortality of marine mammals caused by highly pathogenic influenza virus (H5N1) in South America, bioRxiv, 2023.
Chen, G.W., Gong, Y.N., and Shih, S.R., Influenza A virus plasticity—a temporal analysis of species-associated genomic signatures, J. Formos. Med. Assoc., 2015, vol. 114, no. 5, pp. 456–463.
Zhang, B., Xu, S., Liu, M., et al., The nucleoprotein of influenza A virus inhibits the innate immune response by inducing mitophagy, Autophagy, 2023, vol. 19, no. 7, pp. 1916–1933.
Shi, J., Zeng, X., Cui, P., Yan, C., and Chen, H., Alarming situation of emerging H5 and H7 avian influenza and effective control strategies, Emerging Microbes Infect., 2023, vol. 12, no. 1, p. 2155072.
Adlhoch, C., Fusaro, A., Gonzales, J.L., et al., Avian influenza overview December 2022–March 2023, EFSA J., 2023, vol. 21, no. 3, p. e07917.
Agüero, M., Monne, I., Sánchez, A., et al., Highly pathogenic avian influenza A(H5N1) virus infection in farmed minks, Spain, October 2022, Euro Surveill., 2023, vol. 28, no. 3, p. 2300001.
Smith, G.J., Vijaykrishna, D., Bahl, J., et al., Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic, Nature, 2009, vol. 459, no. 7250, pp. 1122–1125.
Worobey, M., Han, G.Z., and Rambaut, A., Genesis and pathogenesis of the 1918 pandemic H1N1 influenza A virus, Proc. Natl. Acad. Sci. U. S. A., 2014, vol. 111, no. 22, pp. 8107–8112.
Lvov, D.K., Gulyukin, M.I., Zaberezhniy, A.D., and Gulyukin, A.M., Formation of population gene pools of zoonotic viruses, potentially threatening biosafety, Vopr. Virusol., 2020, vol. 65, no. 5, pp. 243–258.
Funding
This work was supported by ongoing institutional funding. No additional grants to carry out or direct this particular research were obtained.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
ETHICS APPROVAL AND CONSENT TO PARTICIPATE
This work does not contain any studies involving human and animal subjects.
CONFLICT OF INTEREST
The authors of this work declare that they have no conflicts of interest.
Additional information
Translated by M. Batrukova
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Chernyshova, A.I., Zhirnov, O.P. Two Phylogenetic Cohorts of the Nucleocapsid Protein NP and Their Correlation with the Host Range of Influenza A Viruses. Dokl Biochem Biophys (2024). https://doi.org/10.1134/S1607672924700789
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1134/S1607672924700789