Abstract
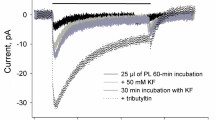
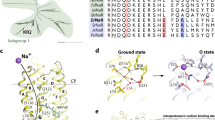
This work provides the first characteristics of the rhodopsin SpaR from Sphingomonas paucimobilis, aerobic bacteria associated with opportunistic infections. The sequence analysis of SpaR has shown that this protein has unusual DTS motif which has never reported in rhodopsins from Proteobacteria. We report that SpaR operates as an outward proton pump at low pH; however, proton pumping is almost absent at neutral and alkaline pH. The photocycle of this rhodopsin in detergent micelles slows down with an increase in pH because of longer Schiff base reprotonation. Our results show that the novel microbial ion transporter SpaR of interest both as an object for basic research of membrane proteins and as a promising optogenetic tool.



Similar content being viewed by others
Change history
24 April 2021
An Erratum to this paper has been published: https://doi.org/10.1134/S1607672921020198
REFERENCES
Gushchin, I. and Gordeliy, V., Microbial rhodopsins, Subcell. Biochem., 2018, vol. 87, pp. 19–56.
Kirpichnikov, M.P. and Ostrovsky, M.A., Optogenetics and vision, Vestn. Ross. Akad. Nauk, 2019, vol. 89, no. 2, pp. 125–130.
Luecke, H., Schobert, B., Richter, H.T., et al., Structure of bacteriorhodopsin at 1.55 A resolution, J. Mol. Biol., 1999, vol. 291, no. 4, pp. 899–911.
Ushakov, A., Grudinin, S., Okhrimenko, I., et al., Knowledge-based prediction model for characterization of microbial rhodopsins for optogenetics, FEBS J., 2016, vol. 283, p. P-03.03.2-014.
Pan, L., Zhou, H., Li, J., et al., Draft genome sequence of Sphingomonas paucimobilis strain LCT-SP1 isolated from the Shenzhou X spacecraft of China, Stand. Genomic Sci., 2016, vol. 11, no. 1, p. 18.
Hardjo Lugito, N.P., Cucunawangsih, and Kurniawan, A., A lethal case of Sphingomonas paucimobilis bacteremia in an immunocompromised patient, Case Rep. Infect. Dis., 2016, vol. 2016, pp. 1–4.
Maragakis, L.L., Chaiwarith, R., Srinivasan, A., et al., Sphingomonas paucimobilis bloodstream infections associated with contaminated intravenous fentanyl1, Emerg. Infect. Dis., 2009, vol. 15, no. 1, pp. 12–18.
Harris, A., Ljumovic, M., Bondar, A.N., et al., A new group of eubacterial light-driven retinal-binding proton pumps with an unusual cytoplasmic proton donor, Biochim. Biophys. Acta—Bioenerg. Elsevier B.V., 2015, vol. 1847, no. 12, pp. 1518–1529.
Stemmer, W.P.C., Crameri, A., Ha, K.D., et al., Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides, Gene, 1995, vol. 164, no. 1, pp. 49–53.
Shevchenko, V., Mager, T., Kovalev, K., et al., Inward H+ pump xenorhodopsin: mechanism and alternative optogenetic approach, Sci. Adv., 2017, vol. 3, no. 9. e1603187.
Bratanov, D., Kovalev, K., Machtens, J., et al., Unique structure and function of viral rhodopsins, Nat. Commun. (Springer US), 2019, vol. 10, no. 1, p. 4939.
Chizhov, I., Chernavskii, D.S., Engelhard, M., et al., Spectrally silent transitions in the bacteriorhodopsin photocycle, Biophys. J. (Elsevier), 1996, vol. 71, no. 5, pp. 2329–2345.
Yoshizawa, S., Kumagai, Y., Kim, H., et al., Functional characterization of flavobacteria rhodopsins reveals a unique class of light-driven chloride pump in bacteria, Proc. Natl. Acad. Sci. U. S. A., 2014, vol. 111, no. 18, pp. 6732–6737.
Gushchin, I., Melnikov, I., Polovinkin, V., et al., Mechanism of transmembrane signaling by sensor histidine kinases, Science, 2017, vol. 356, no. 6342, p. eaah6345.
Farias, M.E., Revale, S., Mancini, E., et al., Genome sequence of Sphingomonas sp. s17, isolated from an alkaline, hyperarsenic, and hypersaline volcano-associated lake at high altitude in the Argentinean Puna, J. Bacteriol., 2011, vol. 193, no. 14, pp. 3686–3687.
Funding
This work was partially supported by the Russian Foundation for Basic Research, project no. 17-00-00167K (KOMFI 17-00-00164, 17-00-00165, 17-00-00166).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflict of interest. This article does not contain any studies involving animals or human participants performed by any of the authors.
AUTHOR CONTRIBUTIONS
N. Maliar, I.S. Okhrimenko, and L.E. Petrovskaya contributed equally to the work.
Additional information
Translated by M. Batrukova
Rights and permissions
About this article
Cite this article
Maliar, N., Okhrimenko, I.S., Petrovskaya, L.E. et al. Novel pH-Sensitive Microbial Rhodopsin from Sphingomonas paucimobilis . Dokl Biochem Biophys 495, 342–346 (2020). https://doi.org/10.1134/S1607672920060162
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1607672920060162