Abstract
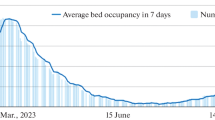
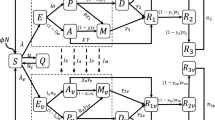
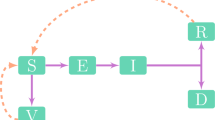
The SARS-COV-2 problem has not been resolved. Waves of the number of recorded infections are continuing locally in the summer of 2023. The threat of repeated epidemic waves will remain relevant for at least another 3 years. In East Asian countries, the number of repeated cases of infection is increasing exponentially due to the immunosuppression caused by long-term replication of SARS-COV-2 in many previously ill people. Repeated COVID illnesses deplete the cell population of CD4 T-lymphocytes. The equation scheme of the SIRS state change models for the COVID epidemic is fundamentally incorrect: each new infection makes a person even more vulnerable. The rate of evolution of the Omicron branch is not decreasing. The new strain BA.2.86 “Pirola” in August carries 36 new mutations compared to the dominant XBB.1.5 “Kraken” in the spring and competes with other newcomers to the zoo of strains: EG.5 Eris, XBB.2.3 Acrux, and FL. 1.5.1 Fornax. SARS-COV-2 differs from related viruses in having a greater rate of renewal of antigens in its S and E proteins. The number of isolated strains is approaching 2000. The key factor in stopping the evolution of the virus is the immunological factor of heterogeneity of epitopes of the population of T-lymphocytes produced in response to infections by past strains. Now, during evolution, the coronavirus needs to find a balance between two tasks: avoiding neutralization by our antibodies and the goal of effectively binding to the cellular receptor ACE2. BA.2.86 avoids neutralization better than XBB and EG, but its transmissibility is not high. “Immuno-evasive” strains are now not so infectious and are still causing regional outbreaks of infections. The diversity of mutational variants of the RNA virus with competition between the evolutionary branches of its strains leads, after attenuation, to new repeated oscillations in the number of infections: the COVID-wave effect. The convergent strain is capable of causing a new outbreak and a global series of slowly fading COVID waves in the winter of 2024. Regional scenarios for COVID-19 epidemics are unusual. The frequency and amplitude of COVID-19 incidence peaks vary significantly across regions, and the author has classified three scenarios. This work provides a model description of the local effects of specific scenarios of wave-like epidemic dynamics. Equations with delay are proposed for complex forms of oscillatory dynamics, supplemented with situational functions of epidemic attenuation. Numerical solutions were obtained for both collapsing and damping oscillations with the possibility of a new outbreak, which made it possible to describe the emerging effect of a single extreme COVID wave in Chile and New York after the activation of infection chains. The model described the occurrence of a sharp peak of infections with a rapid oscillatory decay. The developed model scenarios are very different. Equations with situational functions describe situations of local epidemic waves, a long series of short-term peaks with successively increasing amplitude, as in the epidemic graphs in Japan and Australia, but not in Belgium. Romania in october expects expressive COVID wave.











Similar content being viewed by others
REFERENCES
A. Yu. Perevaryukha, Tech. Phys. 67 (6), 523 (2022). https://doi.org/10.1134/S1063784222070088
A. Yu. Perevaryukha, Tech. Phys. 67 (9), 651 (2022). https://doi.org/10.1134/S1063784222090043
I. V. Trofimova, A. Yu. Perevaryukha, and A. B. Manvelova, Tech. Phys. Lett. 48 (12), 305 (2022). https://doi.org/10.1134/S1063785022110025
V. V. Mikhailov, A. Yu. Perevaryukha, and I. V. Trofimova, Tech. Phys. Lett. 48 (12), 301 (2022). https://doi.org/10.1134/S1063785022110013
A. Yu. Perevaryukha, J. Comput. Syst. Sci. Int. 50, 491 (2011). https://doi.org/10.1134/S1064230711010151
D. Dudakova, V. Anokhin, M. Dudakov, and A. L. Ronzhin, Inf. Autom. 6 (21), 1359 (2022). https://doi.org/10.15622/ia.21.6.10
A. Yu. Perevaryukha, Numer. Anal. Appl. 5, 254 (2012). https://doi.org/10.1134/S199542391203007X
A. Sukhinov, Yu. Belova, A. Nikitina, and V. Sidoryakina, Mathematics 10 (12), 2092 (2022). https://doi.org/10.3390/math10122092
G. Yu. Riznichenko, N. E. Belyaeva, A. N. Diakonova, et al., Biophysics 65 (5), 754 (2020). https://doi.org/10.1134/S0006350920050152
P. V. Veshchev, G. I. Guteneva, and R. S. Mukhanova, Russ. J. Ecol. 43 (2), 142 (2012). https://doi.org/10.1134/S1067413612020154
A. Yu. Perevaryukha, Autom. Control Comput. Sci. 45, 223 (2011). https://doi.org/10.3103/S0146411611040067
T. M. Delorey, C. G. K. Ziegler, G. Heimberg, et al., Nature 595, 107 (2021). https://doi.org/10.1038/s41586-021-03570-8
K. D. Lamb, M. M. Luka, M. Saathoff, R. Orton, et al., SARS-CoV-2’s evolutionary capacity is mostly driven by host antiviral molecules, Preprint Server for Biology: bioRxiv (2023). https://doi.org/10.1101/2023.04.07.536037
C. R. Pothast, R. C. Dijkland, M. Thaler, et al., Elife 11, e82050 (2022). https://doi.org/10.7554/eLife.82050
J. P. A. Ioannidis, S. Cripps, and M. A. Tanner, Int. J. Forecast. 38 (2), 423 (2022). https://doi.org/10.1016/j.ijforecast.2020.08.004
V. Chin, N. I. Samia, R. Marchant, et al., Eur. J. Epidemiol. 35, 733 (2020). https://doi.org/10.1007/s10654-020-00669-6
S. Moein, N. Nickaeen, A. Roointan, et al., Sci. Rep. 11, 4725 (2021). https://doi.org/10.1038/s41598-021-84055-6
A. V. Nikitina, I. A. Lyapunov, and E. A. Dudnikov, Comput. Math. Inf. Technol. 4 (1), 19 (2020). https://doi.org/10.23947/2587-8999-2020-1-1-19-30
R. C. Barnard, N. G. Davies, et al., Nat. Commun. 13, 4879 (2022). https://doi.org/10.1038/s41467-022-32404-y
S. K. Ghosh and S. Ghosh, Sci. Rep. 13, 3610 (2023). https://doi.org/10.1038/s41598-023-30800-y
K. W. Ng, N. Faulkner, G. H. Cornish, et al., Science 370 (6522), 1339 (2020). https://doi.org/10.1126/science.abe1107
A. Yu. Perevaryukha, Cybern. Syst. Anal. 52 (4), 623 (2016). https://doi.org/10.1007/s10559-016-9864-8
G. E. Hutchinson, Ann. N. Y. Acad. Sci. 50 (4), 221 (1948). https://doi.org/10.1111/j.1749-6632.1948.tb39854.x
A. Yu. Kolesov and Yu. S. Kolesov, Tr. MIAN 199, 3 (1993). https://www.mathnet.ru/links/e56394c4bd5687 41eb07ea5ce73abf26/tm1373.pdf
T. Yu. Borisova and A. Yu. Perevaryukha, Tech. Phys. Lett. 48 (7), 251 (2022). https://doi.org/10.1134/S1063785022090012
N. Fabiano and S. N. Radenovic, Mil. Tech. Cour. 69 (1), 1 (2021).
A. V. Shabunin, Izv. Vyssh. Uchebn. Zaved., Prikl. Nelineinaya Din. 30 (6), 717 (2022). https://doi.org/10.18500/0869-6632-003014
M. S. A. Abotaleb and T. A. Makarovskikh, Bull. South Ural State Univ. Ser. Comput. Technol., Autom. Control, Radio Electron. 21 (3), 26 (2021). https://doi.org/10.14529/ctcr210303
A. V. Zaikovskaya, A. V. Gladysheva, M. Yu. Kartashov, et al., Probl. Osobo Opasnykh Infekts., No. 1, 94 (2022). https://doi.org/10.21055/0370-1069-2022-1-94-100
V. I. Vechorko, O. V. Averkov, and A. A. Zimin, Kasdiovaskulyar. Ter. Profil. 21 (6), 89 (2022). https://doi.org/10.15829/1728-8800-2022-3228
S. J. R. da Silva, A. Kohl, L. Pena, and K. Pardee, Rev. Med. Virol. 33 (1), e2373 (2022). https://doi.org/10.1002/rmv.2373
F. E. Juul, H. C. Jodal, I. Barua, et al., Scand. J. Public Health 50, 38 (2022). https://doi.org/10.1177/14034948211047137
N. Brusselaers, D. Steadson, K. Bjorklund, et al., Humanit. Soc. Sci. Commun. 9, 91 (2022). https://doi.org/10.1057/s41599-022-01097-5
Ch. Phetsouphanh, D. R. Darley, D. B. Wilson, et al., Nat. Immunol. 23, 210 (2022). https://doi.org/10.1038/s41590-021-01113-x
H. Oshitani, Nature 605, 589 (2022). https://doi.org/10.1038/d41586-022-01385-9
L. Corey, C. Beyrer, M. S. Cohen, N. L. Michael, T. Bedford, and M. Rolland, N. Engl. J. Med. 385, 562 (2021). https://doi.org/10.1056/NEJMsb210475634347959
E. L. Shrock, R. T. Timms, T. Kula, et al., Science 380 (6640), 798 (2023). https://doi.org/10.1126/science.adc9498
P. V. Markov, M. Ghafari, M. Beer, et al., Nat. Rev. Microbiol. 21, 195 (2023). https://doi.org/10.1038/s41579-023-00878-2
V. V. Mikhailov, A. Yu. Perevaryukha, and Yu. S. Reshetnikov, Inf.-Upravlyayushchie Sist., No. 4 (95), 31 (2018). https://doi.org/10.31799/1684-8853-2018-4-31-38
J. O. Lloyd-Smith, S. J. Schreiber, P. E. Kopp, and W. M. Getz, Nature 438, 355 (2005). https://doi.org/10.1038/nature04153
V. V. Mikhailov, A. V. Spesivtsev, and A. Yu. Perevaryukha, Evaluation of the dynamics of phytomass in the tundra zone using a fuzzy-opportunity approach, in Intelligent Distributed Computing XIII. IDC 2019. Studies in Computational Intelligence, Ed. by I. Kotenko, C. Badica, V. Desnitsky, D. El Baz, and M. Ivanovic (Springer, Cham, 2020), vol. 868, p. 449. https://doi.org/10.1007/978-3-030-32258-8_53
A. Yu. Perevaryukha, Tech. Phys. Lett. 49 (2), 14 (2023). https://doi.org/10.1134/S1063785023010054
A. Yu. Perevaryukha, Biophysics 61 (2), 334 (2016). https://doi.org/10.1134/S0006350916020147
A. Yu. Perevaryukha, Biophysics 66 (2), 327 (2021). https://doi.org/10.1134/S0006350921020160
A. Yu. Perevaryukha, Biophysics 66 (6), 974 (2021). https://doi.org/10.1134/S0006350921060130
A. Yu. Perevaryukha, Tech. Phys. 68 (2), 35 (2023). https://doi.org/10.1134/S106378422301005X
Funding
This study was performed within the framework of the project “Theoretical and Technological Foundations of Digital Transformations of Society and Economy of Russia” of the St. Petersburg Federal Research Center of the Russian Academy of Sciences, grant no. FFZF-2022–0003.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The author declares that she has no conflicts of interest.
Additional information
—For what purpose get your hopes up?
M.Yu. Lermontov, Hero of Our Time
Translated by M. Drozdova
Publisher’s Note.
Pleiades Publishing remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Perevaryukha, A.Y. Scenario Modeling of Regional Epidemics of SARS-COV-2 and Analysis of Immunological Aspects of New Expected COVID Waves. Tech. Phys. 68, 205–217 (2023). https://doi.org/10.1134/S1063784223060038
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063784223060038