Abstract
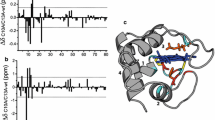
Molecular dynamics simulations of horse heart cytochrome C were performed in an aqueous solution and in a 50 : 50 water–methanol mixture. Three independent 100-ns trajectories were run for each system. It was shown that the addition of methanol significantly affects the mobility of atoms in the cytochrome C molecule in the region from valine 20 to proline 30, which belongs to one of the known cardiolipin-binding sites. During 100-ns MD simulations, the coordination bond between the heme iron atom and the sulfur atom of the nearest methionine is not cleaved, its length is not significantly changed, and the distances from the heme to the residues responsible for fluorescence remain unchanged, as opposed to other studies, in which such changes were observed upon the addition of methanol to cytochrome C.



Similar content being viewed by others
REFERENCES
Z. Zhang, L. Huang, V. M. Shulmeister, et al., Nature 392 (6677), 677 (1998). https://doi.org/10.1038/33612
J. C. Goldstein, N. J. Waterhouse, P. Juin, et al., Nat. Cell. Biol. 2 (3), 156 (2000). https://doi.org/10.1038/35004029
G. W. Bushnell, G. V. Louie, and G. D. Brayer, J. Mol. Biol. 214 (2), 585 (1990). https://doi.org/10.1016/0022-2836(90)90200-6
S. Hirota, M. Ueda, Y. Hayashi, et al., J. Biochem. 152 (6), 521 (2012). https://doi.org/10.1093/jb/mvs098
J. Muenzner and E. V. Pletneva, Chem. Phys. Lipids 179, 57 (2014). https://doi.org/10.1016/j.chemphyslip.2013.11.002
S. Hirota, Y. Hattori, S. Nagao, et al., PNAS 107 (29), 12854 (2010). https://doi.org/10.1073/pnas.1001839107
P. P. Parui, M. S. Deshpande, S. Nagao, et al., Biochem. 52 (48), 8732 (2013). https://doi.org/10.1021/bi400986g
M. S. Deshpande, P. P. Parui, H. Kamikubo, et al., Biochem. 53 (28), 4696 (2014). https://doi.org/10.1021/bi500497s
V. E. Bychkova, A. E. Dujsekina, S. I. Klenin, et al., Biochem. 35 (19), 6058 (1996). https://doi.org/10.1021/bi9522460
R. Grandori, Protein Sci. 11 (3), 453 (2002).
Y. O. Kamatari, T. Konno, M. Kataoka, et al., J. Mol. Biol. 259 (3), 512 (1996). https://doi.org/10.1006/jmbi.1996.0336
D. A. Case, T. E. Cheatham, III, T. Darden, et al., J. Comput. Chem. 26 (16), 1668 (2005). https://doi.org/10.1002/jcc.20290
M. J. Abraham, T. Murtola, R. Schulz, et al., Softwarex 1, 19 (2015). https://doi.org/10.1016/j.softx.2015.06.001
A. W. S. Da Silva and W. F. Vranken, BMC Res. Notes 5 (1), 367 (2012). https://doi.org/10.1186/1756-0500-5-367
D. A. Giammona, Ph. D. Thesis (Davis, University of California, 1984).
J. A. Maier, C. Martinez, K. Kasavajhala, et al., J. Chem. Theory Comput. 11 (8), 3696 (2015). https://doi.org/10.1021/acs.jctc.5b00255
H. J. Berendsen, J. V. Postma, W. F. van Gunsteren, et al., J. Chem. Phys. 81 (8), 3684 (1984). https://doi.org/10.1063/1.448118
M. Parrinello and A. Rahman, J. Chem. Phys. 76 (5), 2662 (1982). https://doi.org/10.1063/1.448118
G. K. Vladimirov, Candidate’s Dissertation in Biology (RNIMU, Moscow, 2018).
V. E. Kagan, H. A. Bayır, N. A. Belikova, et al., Free Radic. Biol. Med. 46 (11), 1439 (2009). https://doi.org/10.1016/j.freeradbiomed.2009.03.004
Funding
The study was supported by the Russian Science Foundation (project no. 19-14-00244; molecular dynamics simulations) and the Ministry of Science and Higher Education of the Russian Federation within the framework of the state assignment for the Federal Scientific Research Centre “Crystallography and Photonics” of the Russian Academy of Sciences (analysis of the results of molecular modeling).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that they have no conflicts of interest.
Additional information
Translated by T. Safonova
Rights and permissions
About this article
Cite this article
Korotkova, P.D., Yurchenko, A.A., Timofeev, V.I. et al. Early Stage of Structural Changes and Molecular Dynamics of Cytochrome C in an Aqueous Solution Caused by the Addition of Methanol. Crystallogr. Rep. 67, 229–232 (2022). https://doi.org/10.1134/S1063774522020079
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063774522020079