Abstract
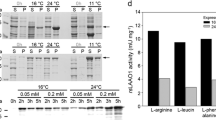
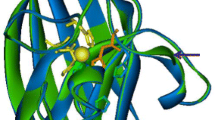
Protein acetylation is one of the most common post-translational modifications. Many acetylated proteins in Magnaporthe oryzae play key roles in vegetative growth and pathogenicity. MoDabb1 from M. oryzae was also identified to be an acetylated protein, containing a Dabb domain with unknown function. To elucidate the function of this protein and the effect of acetylation on this protein, a native and selenomethionine-substituted MoDabb1 were expressed in Escherichia coli and purified to homogeneity. Crystals were obtained using sitting-drop vapour-diffusion method. Crystals of native and selenomethionine-substituted protein were diffracted to a resolution of 1.74 and 1.98 Å and both belonged to the sp. gr. P42212. Matthews coefficient analysis indicated two molecules in an asymmetric unit with a Vm value of 2.41 and a corresponding solvent content of 49.03%.



Similar content being viewed by others
REFERENCES
A. Drazic, L. M. Myklebust, R. Ree, et al., Biochim. Biophys. Acta 1864, 1372 (2016).
Q. Wang, Y. Zhang, C. Yang, et al., Science 327, 1004 (2010).
S. Nambi, K. Gupta, M. Bhattacharyya, et al., J. Biol. Chem. 288, 14114 (2013).
S. Zhao, W. Xu, W. Jiang, et al., Science 327, 1000 (2010).
K. L. Guan and Y. Xiong, Trends Biochem. Sci. 36, 108 (2011).
C. Choudhary, C. Kumar, F. Gnad, et al., Science 325, 834 (2009).
J. Ren, Y. Sang, Y. Tan, et al., PLOS Pathog. 12, e1005458 (2016).
D. Li, B. Lv, L. Tan, et al., Sci. Rep. 6, 29897 (2016).
P. Henriksen, S. A. Wagner, B. T. Weinert, et al., Mol. Cell Proteomics 11, 1510 (2012).
G. Wang, L. Guo, W. Liang, et al., AMB Express 7, 94 (2017).
B. Lv, Q. Yang, D. Li, et al., Sci. Rep. 6, 29313 (2016).
S. Zhou, Q. Yang, C. Yin, et al., BMC Genomics 17, 1019 (2016).
R. Dean, J. A. Van Kan, Z. A. Pretorius, et al., Mol. Plant Pathol. 13, 414 (2012).
R. A. Dean, N. J. Talbot, D. J. Ebbole, et al., Nature 434, 980 (2005).
X. Sun, Z. Li, H. Liu, et al., Sci. Rep. 7, 15316 (2017).
Z. Qi, Q. Wang, X. Dou, et al., Mol. Plant Pathol. 13, 677 (2012).
B. C. Osmond, C. A. Specht, and P. W. Robbins, Proc. Natl. Acad. Sci. U.S.A. 96, 11206 (1999).
Y. Li, X. Yan, H. Wang, et al., Mol. Plant Microbe Interact. 23, 317 (2010).
R. N. Patkar, M. Ramos-Pamplona, A. P. Gupta, et al., Mol. Microbiol. 86, 1345 (2012).
S. F. Altschul, T. L. Madden, A. A. Schäffer, et al., Nucleic Acids Res. 25, 3389 (1997).
R. Gu, S. Fonseca, L. G. Puskas, et al., Tree Physiol. 24, 265 (2004).
A. Walker, J. Taylor, D. Rowe, et al., Plasmid 59, 155 (2008).
Z. Otwinowski and W. Minor, in Methods in Enzymology (Academic Press, 1997), p. 307.
M. D. Winn, C. C. Ballard, K. D. Cowtan, et al., Acta Crystallogr. D: Biol. Crystallogr. 67, 235 (2011).
B. Heras and J. L. Martin, Acta Crystallogr. D: Biol. Crystallogr. 61, 1173 (2005).
B. W. Matthews, J. Mol. Biol. 33, 491 (1968).
D. E. Kim, D. Chivian, and D. Baker, Nucleic Acids Res. 32, W526 (2004).
L. A. Kelley, S. Mezulis, C. M. Yates, et al., Nat. Protoc. 10, 845 (2015).
ACKNOWLEDGMENTS
We are grateful to Dr. Arie Geerlof at EMBO for kindly supplying the expression vector pHAT2. We also thank all the staffs from BL18U1 and BL19U1 beamline of National Facility for Protein Science Shanghai (NFPS) at Shanghai Synchrotron Radiation Facility (SSRF) for help with crystal screening and data collection. This research was supported by the National Key Basic Research and Development Program (no. 2017YFD0201705), the National Natural Science Foundation of China (NSFC) (nos. 31471735 and 31772112), and the Taishan Scholar Construction Foundation of Shandong Province (no. 6631114314).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhao, Y., Chi, M., Zhang, X. et al. Expression, Purification, Crystallization and X-Ray Crystallographic Analysis of MoDabb1 from Magnaporthe oryzae. Crystallogr. Rep. 64, 1112–1116 (2019). https://doi.org/10.1134/S1063774519070307
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S1063774519070307