Abstract
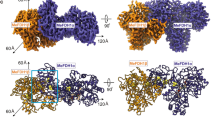
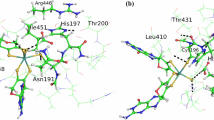
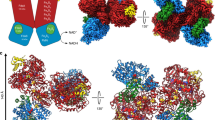
Formate dehydrogenase (FDH) from the methylotrophic bacterium Pseudomonas sp. 101 catalyzes oxidation of formate to NI2 with the coupled reduction of nicotinamide adenine dinucleotide (NAD+). The three-dimensional structures of the apo form (the free enzyme) and the holo form (the ternary FDH-NAD+-azide complex) of FDH have been established earlier. In the present study, the structures of FDH complexes with formate are solved at 2.19 and 2.28 Å resolution by the molecular replacement method and refined to the R factors of 22.3 and 20.5%, respectively. Both crystal structures contain four protein molecules per asymmetric unit. These molecules form two dimers identical to the dimer of the apo form of FDH. Two possible formatebinding sites are found in the active site of the FDH structure. In the complexes the sulfur atom of residue Cys354 exists in the oxidized state.
Similar content being viewed by others
References
V. S. Lamzin, Z. Dauter, and V. O. Popov, J. Mol. Biol. 236, 759 (1994).
E. V. Filippova, K. M. Polyakov, and T. V. Tikhonova, Kristallografiya 5(5), 857 (2005) [Crystallogr. Rep. 50, 796 (2005)].
Z. Otwinowski and W. Minor, Methods Enzymol. 276, 307 (1997).
A. A. Vagin and A. Teplyakov, J. Appl. Crystallogr. 30, 1022 (1997).
G. N. Murshudov, A. A. Vagin, and E. J. Dodson, Acta Crystallogr. D 53, 240 (1997).
T. A. Jones, J.-Y. Zou, S. W. Cowan, and M. Kjeldgaard, Acta Cristallogr. A 47, 110 (1991).
A. Roussel and C. Cambillau, Silicon Graphics (Silicon Graphics, Mountain View, 1991), p. 81.
SERC (UK) Collaborative Computing Project No. 4. Acta. Crystallogr. A. 46, 585 (1994).
A. S. Kutzenko, V. S. Lamzin, and V. O. Popov, FEBS Lett. 423, 105 (1998).
T. B. Ustinnikova, V. O. Popov, and Ts. A. Egorov, Bioorg. Khim. 14, 905 (1988).
V. I. Tishkov, A. G. Galkin, G. N. Marchenko, et al., Biochem. Biophys. Res. Commun. 192, 976 (1993).
E. R. Odintseva, A. S. Popova, A. M. Rozhkova, and V. I. Tishkov, Vestn. Mosk. Univ., Ser. Khim. 43, 356 (2002).
Author information
Authors and Affiliations
Additional information
Original Russian Text © E.V. Filippova, K.M. Polyakov, T.V. Tikhonova, T. N. Stekhanova, K.M. Boiko, I.G. Sadykhov, V.I. Tishkov, V.O. Popov, N. Labru, 2006, published in Kristallografiya, 2006, Vol. 51, No. 4, pp. 667–671.
Rights and permissions
About this article
Cite this article
Filippova, E.V., Polyakov, K.M., Tikhonova, T.V. et al. Crystal structures of complexes of NAD+-dependent formate dehydrogenase from methylotrophic bacterium Pseudomonas sp. 101 with formate. Crystallogr. Rep. 51, 627–631 (2006). https://doi.org/10.1134/S1063774506040146
Received:
Issue Date:
DOI: https://doi.org/10.1134/S1063774506040146