Abstract
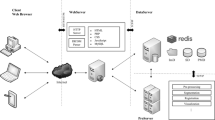
Image analysis plays an important role both in medical diagnostics and in biology. The main reasons that prevent the creation of objective and reliable methods of analysis of biomedical images are the high variability and heterogeneity of the biological material, distortion introduced by the experimental procedures, and the large size of the images. This paper presents preliminary results on creating a system called Ter-aPro, which combines a platform for image processing (ProStack) and a raster data storage system (rasdaman). This integrated system can be used in a cloud environment, providing access to the methods of visualization, analysis, and processing of a large amount of images through the Internet. Such an approach increases the speed and quality of image understanding and softens the limitations imposed by other systems. The system allows us to view uploaded images in the browser without having to install additional software on any device connected to the Internet, such as tablet computers and smartphones. This paper presents the preliminary results of processing biomedical images.
Similar content being viewed by others
References
P. Widener, M. Barrick, J. Pullikottil, P. Bridges, and A. Maccabe, “A framework for out-ofband processing in large-scale data flows,” in Proc. 2007 Int. Conf. on Grid Computing (Grid 2007) (Austin, 2007).
U. Bick and H. Lenzen, “PACS: the silent revolution,” Europ. Radiol. 9(6), 1152–1160 (1999).
I. Goldberg, C. Allan, J.-M. Burel, D. Creager, A. Falconi, H. Hochheiser, J. Johnston, J. Mellen, P. Sorger, and J. Swedlow, “The open microscopy environment (OME) data model and XML file: Open tools for informatics and quantitative analysis in biological imaging,” Genome Biol. 6, R47 (2005).
P. M. Kuzmak and R. E. Dayhoff, “Integration of imaging functionality into the healthcare enterprise using DICOM,” J. Digital Imag. 11(3), 67–70 (1998).
I. B. Gurevich, O. Salvetti, and Y. O. Trusova, “Fundamental concepts and elements of image analysis ontology,” Pattern Recogn. Image Anal. 19(4), 603–611 (2009).
R. C. Gonzalez and R. E. Woods, Digital Image Processing, 2nd ed. (Prentice Hall, Upper Saddle River, N.J., 2002).
K. N. Kozlov, A. S. Pisarev, and M. G. Samsonova, PC Software Certificate no. 2009611452 (March 16, 2009). http://www.prostack.ru
G. Bradski, “The OpenCV Library,” Dr. Dobb’s J. Software Tools (2000).
P. Baumann, “On the management of multidimensional discrete data,” VLDB J. 4, 401–444 (1994).
T. Kauppi, V. Kalesnykiene, J.-K. Kamarainen, L. Lensu, I. Sorri, A. Raninen, R. Voutilainen, H. Uusitalo, H. Kalviainen, and J. Pietila, “DIARETDB1 diabetic retinopathy database and evaluation protocol,” Tech. Rep. (2008). http://www.it.lut.fi/project/imageret
S. Ravishankar, A. Jain, and A. Mittal, in Proc. 2009 Int. Conf. on Grid Computing (Grid 2009), CVPR (Banff, 2009), pp. 210–217.
J. Jaeger, S. Surkova, M. Blagov, H. Janssens, D. Kosman, K. N. Kozlov, Manu, E. Myasnikova, C. E. Vanario-Alonso, M. Samsonova, D. H. Sharp, and J. Reinitz, “Dynamic control of positional information in the early Drosophila embryo,” Nature 430, 368–371 (2004).
J. Jaeger, M. Blagov, D. Kosman, K. N. Kozlov, Manu, E. Myasnikova, S. Surkova, C. E. Vanario-Alonso, M. Samsonova, D. H. Sharp, and J. Reinitz, “Dynamical analysis of regulatory interactions in the gap gene system of Drosophila melanogaster,” Genetics 167, 1721–1737 (2004).
V. V. Gursky, J. Jaeger, K. N. Kozlov, J. Reinitz, and A. M. Samsonov, “Pattern formation and nuclear divisions are uncoupled in Drosophila segmentation: Comparison of spatially discrete and continuous models,” Phys. D 197, 286–302 (2004).
M. G. Samsonova, V. V. Gurskii, K. N. Kozlov, and A. M. Samsonov, “System approach for organisms researching,” Nauch.-Tekhn. Vedomosti St. Petersburg Gos. Tekhn. Univ. 2, 222–234 (2006).
Manu, S. Surkova, A. V. Spirov, V. V. Gursky, H. Janssens, A.-R. Kim, O. Radulescu, C. E. Vanario-Alonso, D. H. Sharp, M. Samsonova, and J. Reinitz, “Canalization of gene expression in the Drosophila blastoderm by dynamical attractors,” Plos Biol. 7, e1000049 (2009).
Manu, S. Surkova, A. V. Spirov, V. V. Gursky, H. Janssens, A. R. Kim, O. Radulescu, C. E. Vanario-Alonso, D. H. Sharp, M. Samsonova, and J. Reinitz, “Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors,” PLoS Comput. Biol. 5, e1000303 (2009).
S. Surkova, E. Myasnikova, H. Janssens, K. Kozlov, A. A. Samsonova, J. Reinitz, and M. Samsonova, “Pipeline for acquisition of quantitative data on segmentation gene expression from confocal images,” Fly (Austin) 2, 58–66 (2008).
A. Pisarev, E. Poustelnikova, M. Samsonova, and J. Reinitz, “FlyEx, the quantitative atlas on segmentation gene expression at cellular resolution,” Nucl. Acids Res. 37, D560–D566 (2009).
Author information
Authors and Affiliations
Corresponding author
Additional information
Peter Baumann is Professor of Computer Science at the University of Jacobs (Germany, Bremen), head of various national and international research and industrial projects since 1996, including in the 6th and 7th EU Framework Programmes. During the EU FP6 ESTEDI project, the group of Peter Baumann successfully collaborated with the group of Prof. Maria Georgievna Samsonova.
Peter Baumann is a world expert in the field of large-scale image services with more than 95 publications in reviewed journals, innovative work in the field of scientific databases, in which he explored various aspects of support for data arrays in DBMS, including mathematical modeling, development of a query language, architecture, optimization, implementation, and application in remote probing, the study of climate, oceanography, astrophysics, brain research, analysis gene activity, education, etc. He is the chief architect of rasdaman, which has been used for the past five years to support objects weighing dozens of terabytes.
Since 2002, Peter Baumann has been actively involved in standardization as an expert. He is a cochairman of the working groups on processing raster data in the Open Geospatial Consortium (OGC), which engages in standardization in the field of geo-services in collaboration with ISO, OASIS-Open, and W3C. He currently is the editor of nine specifications that are adopted as a standard (WCPS) or are in process of being approved in July 2010 (WCS 2.0).
Jost Waldmann works on a project at the Jacobs University under the supervision of Peter Baumann.
Konstantin N. Kozlov graduated from the Department of Applied Mathematics, Faculty of Physics and Mechanics, St. Petersburg State Polytechnic University in 2000. His experience of scientific work is more than 10 years. He has worked at the Department of Computational Biology, Center for Advanced Studies, since 2001. Dr. Kozlov is a specialist in applied mathematical methods and information technologies, and conducts scientific research in the field of biomedical image processing and mathematical modeling of biological systems, is a coauthor of more than 35 works published in leading journals. He is member of international conferences, an invited speaker at the “Evolution, Systems Biology, and Supercomputing in Bioinformatics” International School Seminar at the International Conference on Bioinformatics of Genome Regulation and Structure (BGRS), 2006.
Mariya G. Samsonova is a Professor and Doctor of Biological Sciences. She is Head of the Department of Computational Biology at the Center for Advanced Studies. Prof. Mariya Samsonova has headed many Russian and international research projects. She has extensive experience in the creation of consortia, writing projects, and project management. Samsonova is the author of over 70 publications in reviewed journals, a member of the Editorial Board of the Journal of Integrative Bioinformatics and Mathematical Biology and Bioinformatics, invited speaker at many international conferences on bioinformatics and system biology. She received an award for best paper at the ISMB99 conference. Samsonova is an expert in developing methods of extracting quantitative information on the activity of the genes from images, in database development, integration of distributed databases, programs, and services, as well as natural language interfaces. The group of Prof. Mariya Samsonova has developed innovative methods and software for system biology that are widely used by the scientific community. Among them are the ProStack image processing package, BREReA software for registration and removal of background signal from gene activity pictures, and the FlyEx database (http://urchin.spbcas.ru/flyex/).
Rights and permissions
About this article
Cite this article
Kozlov, K.N., Baumann, P., Waldmann, J. et al. TeraPro, a system for processing large biomedical images. Pattern Recognit. Image Anal. 23, 488–497 (2013). https://doi.org/10.1134/S105466181304007X
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S105466181304007X